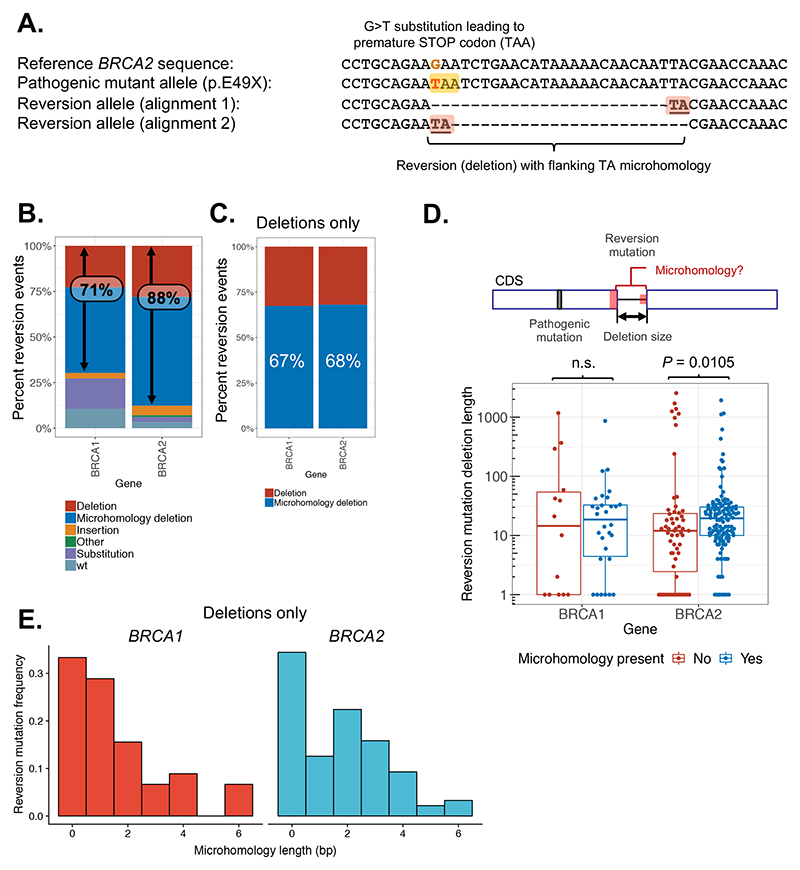
Figure 3. Microhomology usage in reversion mutations.
A. Example of a reversion mutation in BRCA2 associated with microhomology (patient 201 from Cruz et al. (52)). The pathogenic G>T substitution mutation (BRCA2 c.145G>T) introduces a premature stop codon (TAA) as shown. The reversion mutation (c.145_168del24) is an in-frame deletion removing the mutated codon (shown in two different alignments). The existence of microhomology at this deletion is illustrated by the ambiguous alignment of the two nucleotides (TA) flanking it – these could be aligned equally well at either end as illustrated. B. Bar chart of reversion events classified by type. Reversions occurring via deletion are more frequent in BRCA2 (88%) than in BRCA1 (71%). C. Within deletion mutations, the use of microhomology occurs at a similar frequency in BRCA1 and BRCA2. Reversion mutations are plotted as in (B) for deletions only. D. Deletion sizes are generally larger in BRCA2 reversions (P = 0.0105, Wilcoxon rank sum test) with evidence of microhomology use. Total length of deleted sequence is shown for each reversion event, broken down by gene and presence of microhomology. E. BRCA2 reversions use longer lengths of microhomology compared to BRCA1. Frequency distribution of length of microhomology used in BRCA1 (red, left – mode 1 bp) compared with BRCA2 (blue, right – mode 2 bp) plotted for all secondary deletions.