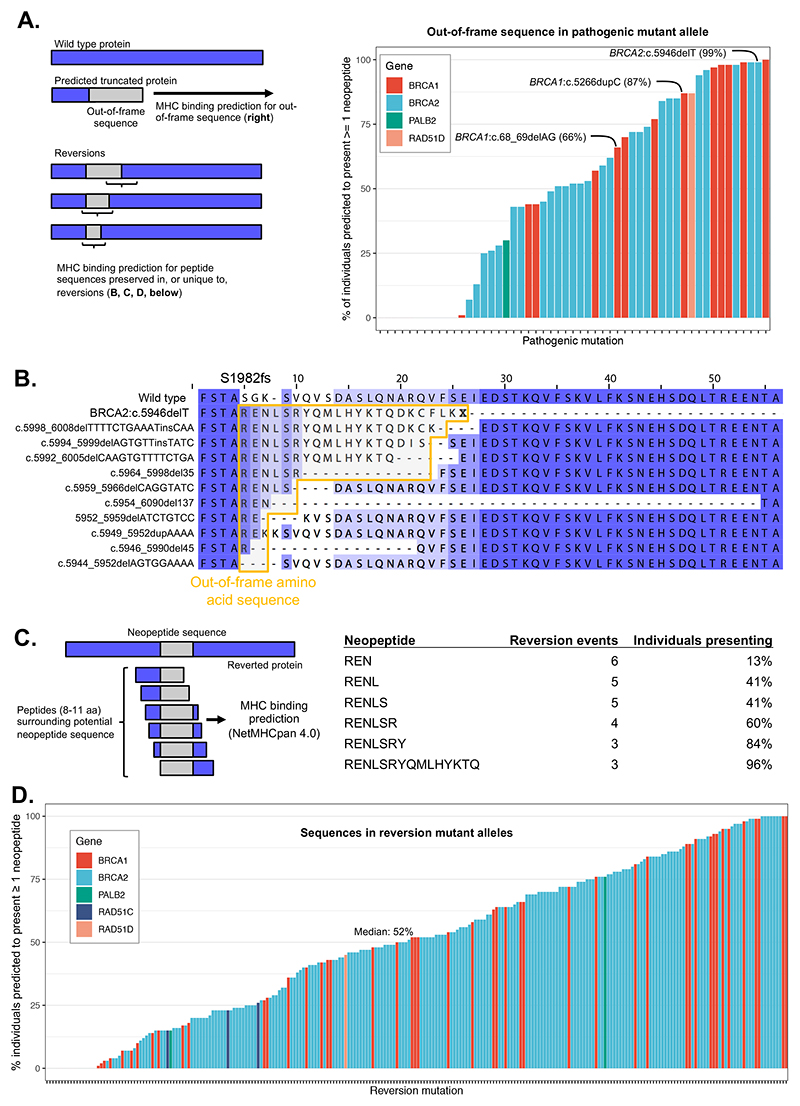
Figure 4. Prediction of HLA-mediated antigen presentation of reversion peptides.
A. Percentage of individuals predicted to present at least one neopeptide from out-of-frame sequence associated with the listed pathogenic deletion mutations. This sequence will be shared with reversion mutations to some extent depending on the position of the reversion relative to the pathogenic mutation. Common founder mutations are highlighted. B. Predicted amino acid sequences from BRCA2:c.5946delT [c.6174delT] reversion events showing retention of out-of-frame sequence in many reversion alleles. The predicted protein sequence for each reversion observed for BRCA2:c.5946delT is shown compared to the wild-type (top) and predicted truncated c.5946delT protein sequence (second row). Sequences deriving from translation of out-of-frame coding sequence are shown in the yellow box. Amino acids are shaded based on their alignment to the wild type sequence. C. Computational prediction of HLA (HLA-A, HLA-B, HLA-C) presentation of out-of-frame protein sequences from BRCA2 c.5946delT downstream reversions. Presentation likelihood calculated using NetMHCpan 4.0. The table shows the proportion of individuals in a set of 1,261 from the 1000 genomes project that have an HLA type predicted to present (%rank < 0.5) at least one neopeptide (length 8 to 11) associated with the indicated out-of-frame sequence (note that such neopeptides can include one or more WT amino acids upstream of the out-of-frame sequence). D. Percentage of individuals predicted to present at least one neopeptide for reverted protein sequences from all published cases of reversion mutations that encode neopeptides.