Abstract
A growing appreciation of the importance of cellular metabolism together with recent revelations concerning the extent of cell-cell heterogeneity demand performing metabolic characterization of individual cells. We present SpaceM, an open-source method for in situ single-cell metabolomics that detects >100 metabolites from >1,000 individual cells/hour together with a fluorescence based read-out and morpho-spatial features. We validated SpaceM by predicting the cell types of co-cultured human epithelial cells and mouse fibroblasts. We used SpaceM to show that stimulating human hepatocytes with fatty acids led to the emergence of two co-existing subpopulations outlined by distinct cellular metabolic states. Inducing inflammation with the cytokine IL-17A perturbs the balance of these states in a process dependent on NF-κB signalling. The metabolic-state markers were reproduced in a pre-clinical in vivo murine model of non-alcoholic steatohepatitis. We anticipate SpaceM to be broadly applicable for investigations of diverse cellular models and to democratize single-cell metabolomics.
Introduction
Organisms are composed of cells of various types, in different states, and functions. A key component is the intracellular metabolome, a repertoire of metabolites and lipids involved in virtually all cellular processes. Recent discoveries transformed our understanding of metabolites and lipids beyond being metabolic building blocks and shed light on their roles in signaling, epigenome regulation, immunity, inflammation, and cancer1,2. The importance of the metabolome, and the emerging capacities of metabolomics makes metabolome profiling an attractive approach to read out the state of the organism, organ, and cells3,4.
In parallel, single-cell technologies have revolutionized biology by highlighting the extent of cellular heterogeneity within tissues5 and even among monoclonal cells6–8. However, investigating metabolism at the single-cell level in situ was out of reach until recently9. Over the past years, pioneered by 10,11, the introduction of single-cell metabolomics methods demonstrated the feasibility of characterizing metabolic heterogeneity and discriminating cell types9,12–15. Certainly, their relatively low throughput, requirements for custom mass spectrometry instrumentation, challenges in analysis of cells cultured at a high density, and the lack of computational methods for downstream analyses, altogether led to a limited accessibility of single-cell metabolomics.
Here, we aim to democratize single-cell metabolomics for cultured cells by introducing an open-source method, SpaceM, along with a toolbox of supplemental methods for metabolite intensity normalization and filtering, quality control, downstream data analysis, and interpretation. SpaceM is compatible with conventional cell culture practices, and can be applied to cells at high confluency. Importantly, our method does not require a custom mass spectrometry setup but rather uses commercially-available Matrix Assisted Laser Desorption Ionization (MALDI)-imaging mass spectrometry and light microscopy. The cornerstone of SpaceM is the precise estimation of which cell parts were locally ablated by the laser, achieved by co-registering microscopy images of cells with microscopy images of MALDI laser ablation marks with a subcellular precision.
Ultimately, SpaceM generates a spatio-molecular matrix for each cell containing a normalized metabolic profile and microscopy-derived phenotypic properties such as cell fluorescence intensities and morpho-spatial features. SpaceM detects >100 metabolites and lipids from >1,000 cells per hour, demonstrating high sensitivity and throughput. The single-cell resolution of SpaceM was validated using our proposed model of co-cultured human epithelial cells (HeLa) and mouse fibroblast cells (NIH3T3) where the cell types were predicted with an accuracy of 91.3% solely based on the single-cell metabolomes.
Using SpaceM, we characterized metabolic states within an isogenic cell population of human differentiated hepatocytes, dHepaRGs, an established in vitro model of Non-Alcoholic Fatty Liver Disease (NAFLD) and Non-Alcoholic Steatohepatitis (NASH)16,17, characterized by a profound and heterogeneous lipid metabolism remodelling18. SpaceM provided single-cell profiles from 29,738 hepatocytes representing intensities of >700 lipids from various classes, including neutral glycerolipids (e.g. triglycerides), glycerophospholipids and sphingolipids. Among hepatocytes stimulated with fatty acids, we discovered a subpopulation with distinctly altered lipid profiles. These hepatocytes consistently compose 24% of the population and are characterized by an aberrant accumulation of lipid droplets and neutral lipids characteristic of a metabolic state called steatosis. We explored how inflammation affects hepatocyte heterogeneity by further challenging them with the cytokine IL-17A, a known player in NASH-associated inflammation19. This shifted almost all hepatocytes (93%) into the steatotic metabolic state and impacted metabolic cell-cell heterogeneity. Furthermore, SpaceM revealed that inhibiting canonical NF-κB signalling alters the relative cell distribution within metabolic states, indicating a potential role for this pathway in regulating hepatocyte metabolic heterogeneity under inflammation-induced conditions. Our metabolic state markers are in line with previous literature and were structurally and quantitatively validated using bulk lipidomics of an in vivo NASH murine model20. This highlights the relevance of the dHepaRG in vitro NAFLD/NASH model and the capacity of SpaceM to discover and characterize biologically-relevant metabolic states. Overall, considering the demonstrated potential of SpaceM to reveal and characterize disease-relevant metabolic states, as well as the accessible nature of the method, we expect to democratize single-cell metabolomics of diverse cell cultures and co-cultures.
Results
The SpaceM method
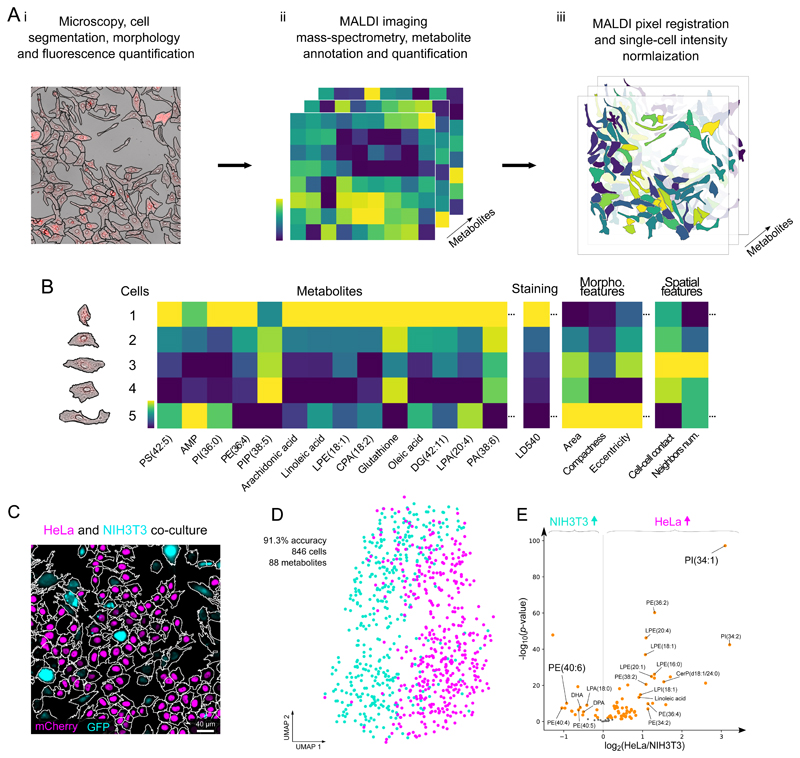
A key challenge of MALDI-based single-cell metabolomics is the assignment of metabolite intensities to individual cells. To address this, we propose SpaceM as a method for single-cell metabolomics of cultured cells. SpaceM integrates MALDI-imaging with light microscopy followed by image segmentation and registration (Figure 1A, Supp. Fig. 1).
Figure 1. The SpaceM workflow and validation.
SpaceM integrates light microscopy and MALDI-imaging mass spectrometry to provide a metabolic profile obtained in situ for each cell. Segmented microscopy images help delineate cells and quantify their morphometric properties, spatial organization, and fluorescence. The cells are further subjected to MALDI-imaging mass spectrometry and metabolite annotation which reveals their metabolomes, followed by normalization. B: SpaceM provides a single-cell spatio-molecular matrix that integrates the metabolic profiles and other information obtained using microscopy. C: We validated SpaceM by predicting cell types of co-cultured human HeLa epithelial cells (H1B-mCherry, magenta) and mouse NIH3T3 fibroblasts (GFP, cyan). Representative image from 2 independent experiments. D: UMAP visualization of 846 co-cultured cells (HeLa, magenta; NIH3T3, cyan) using intensities of 88 metabolites. Unsupervised Leiden clustering applied to the metabolic profiles classified both cell types with an accuracy of 91.3% (see also Figure S7). E: Volcano plot (log2 of the fold change HeLa/NIH3T3 vs. -log10 of the two-tailed independent t-test p-value) showing differential properties of the 88 detected metabolites.
For SpaceM, cells need to be cultured on a microscopy-compatible substrate, followed by fixation, fluorescence staining and drying. SpaceM starts with the acquisition of microscopy images, called pre-MALDI. Cell segmentation of pre-MALDI images provides a broad panel of phenotypic information including fluorescent intensities and spatio-morphological properties of individual cells (Figure 1A i). Next, the sample is sprayed with a MALDI matrix salt and MALDI-imaging mass spectrometry is performed for untargeted detection of molecules, including metabolites and lipids21 (Figure 1A ii).
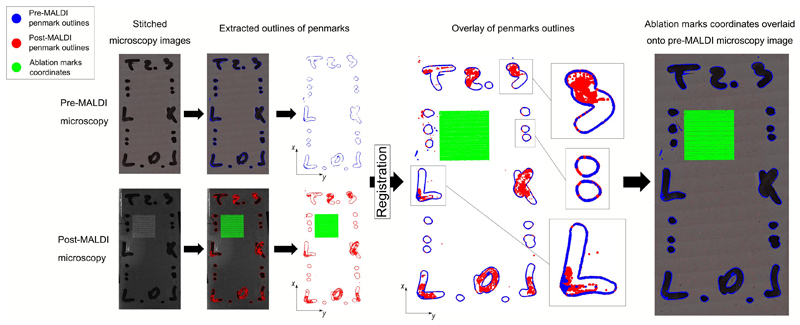
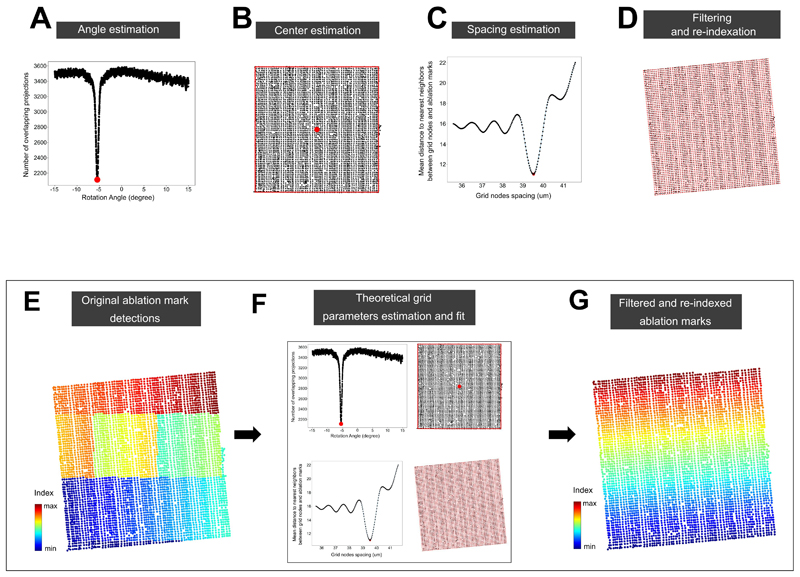
The next steps of SpaceM aim to detect the sampled cell areas. The MALDI laser leaves discernable ablation marks in the overlaying matrix which can be used to identify the areas contributing to metabolite intensities. However, the opacity of the MALDI-matrix prevents simultaneous inspection of both cells and ablation marks. Therefore, the pre-MALDI image is co-registered with a second round of microscopy images, called post-MALDI, from which ablation marks are segmented (Extended Data Fig 1). Although methods to integrate MALDI-imaging data and microscopy exist, their registration accuracies are worse than 1 μm. A prominent example includes registration of MALDI-imaging data of tissue sections with histological images by detecting ablation marks in autofluorescence microscopy images with the reported registration accuracy of 2.2 μm 22. While such accuracy is suitable for tissue section analysis, the aim of SpaceM is to precisely evaluate the ablated parts of single cells. For this reason we developed a high-precision image registration that employs pen marks drawn at the edges of the cell culture which provided over 400,000 fiducial points (Extended Data Fig 2). We estimated that at least 3,000 fiducials are needed to achieve a sub-micron registration accuracy (Supp. Fig. 2), a prerequisite for estimating metabolite intensities at the single-cell level, given that in our study cell sizes range from 37μm to 59μm (25th/75th percentiles, 37μm to 49μm for the dHepaRGs, 39μm to 59μm for the NIH3T3 and 30μm to 39μm for the HeLa cells) (Supp. Fig. 3). Finally, the ablation marks are re-indexed (Extended Data Fig 3) to couple MALDI data with microscopy.
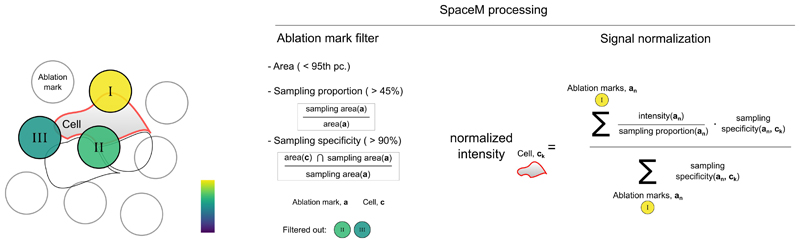
To construct single-cell metabolic profiles from MALDI measurements, we developed an approach accounting for the overlap between cells and ablation marks (Figure 1A iii) to compensate for differences in cell sampling and filter out ambiguous ablation marks sampling multiple cells (Extended Data Fig 4). Ultimately, SpaceM provides a single-cell spatio-molecular matrix with a multiplex readout for each individual cell quantified in its native spatial context. This readout comprises an untargeted metabolic profile, fluorescence intensities and spatio-morphological features (Figure 1B). This information can be used to link metabolism and phenotype at the single-cell level.
Validating SpaceM by predicting cell types with single-cell resolution
To validate SpaceM, we predicted the cell types of co-cultured human HeLa cells and mouse NIH3T3 fibroblasts, constitutively expressing H2B-mCherry and GFP respectively, solely based on their single-cell metabolic profiles (Figure 1C–E). To confirm the prediction, cell types were determined using a linear boundary between mCherry and GFP intensities (Supp. Fig. 4A). SpaceM provided metabolic profiles of 88 metabolites for 445 HeLa and 400 NIH3T3 co-cultured cells (Figure 1C). Uniform Manifold Approximation and Projection (UMAP) visualization based on metabolite intensities of two replicates reproducibly grouped cells by their type (Supp. Fig. 4B). Metabolite intensities were also consistent across replicates, indicating low analytical variability (Supp. Fig. 4C). Integration of the replicates preserved the relative organisation of cell types in UMAP (Supp. Fig. 4D). The linear discriminant analysis performed on the metabolic profiles resulted in the classification of HeLa from NIH cells with an accuracy of 91.3% (Figure 1D, Supp. Fig. 4E). The cell type markers include phosphatidylinositols PI(34:1) and PI(34:2) for HeLa cells and phosphoethanolamine PE(40:6) for NIH3T3 (Figure 1E). Importantly, cells surrounded by the other cell type were correctly classified, demonstrating the single-cell precision of SpaceM (Supp. Fig. 4F–G). We investigated the impact of co-sampling neighboring cells and diffusible metabolites on the cell type classification. For this, we took advantage of the unbiased sampling of SpaceM and constructed the diffusion profile of each detected metabolite (Supp. Fig. 5). Removing all co-sampling ablation marks and discarding the diffused metabolites did not affect the cell type classification accuracy (Supp. Fig. 6A). The classification accuracy was optimal when including co-sampling ablation marks that share at least 90% of their sampling area with a cell of interest (Supp. Fig. 6B–D). Metabolic markers were validated by using individual mono-cultures of each cell type analyzed with both SpaceM and LC-MS/MS (Supp. Fig. 7). Overall, the fold change trends of the cell type markers were validated for 85.2% of the detected metabolites (Supp. Fig. 7A, Supp. Fig. 7B, Supp. Fig. 7D). Using LC-MS/MS, a total of 19 lipids were detected in common with SpaceM; of which 15 lipids showed the same trends (Supp. Fig. 7C). We found the relative metabolite intensities of cell-type-markers to be highly consistent between culture conditions although the NIH3T3 cells had higher intensities when co-cultured with HeLa cells, potentially indicating a metabolic adaptation (Supp. Fig. 7E–F).
SpaceM identifies a steatotic subpopulation of human hepatocytes
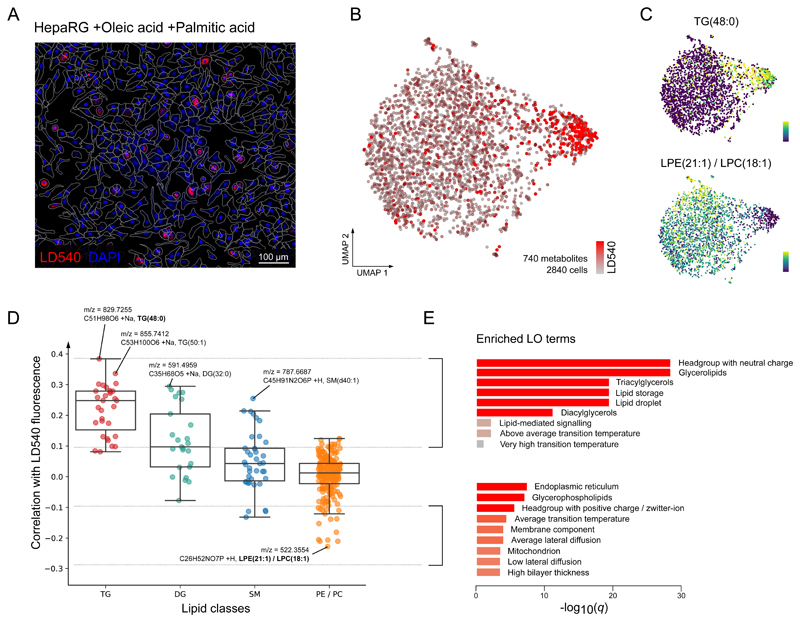
Beyond characterizing cell types, we used SpaceM to identify metabolic states within an isogenic cell line of differentiated human hepatocytes (dHepaRG) upon stimulation with fatty acids (Figure 2A). This model is of particular relevance to study the onset of NAFLD, in which hepatocytes undergo lipid metabolism remodeling and accumulate intracellular lipid droplets (LDs), known as steatosis. NAFLD, in turn, can progress towards NASH, characterized by systemic and hepatic inflammation19. The known high heterogeneity of LD accumulation in hepatocytes 18 (also illustrated in Figure 2A) reinforces the need for single-cell methods.
Figure 2. SpaceM identifies a steatotic subpopulation in lipid stimulated human hepatocytes.
A: Microscopy images of the human dHepaRG hepatocytes stimulated with oleic and palmitic fatty acids illustrating their heterogeneity in lipid droplets accumulation; red: LD540 staining for lipid droplets; blue: Hoechst staining for cell nuclei. Representative image from 4 independent experiments. B: UMAP visualization of single-cell metabolic profiles (740 metabolites) of 2,840 single cells revealed two co-existing subpopulations with statistically different lipid droplet levels (see also Figure S9). C: UMAP plots showing single-cell intensities of metabolites most associated with the lipid droplet accumulation: triglyceride TG(48:0), Spearman correlation r=0.38 (two-tailed test p-value=2.04e-100, ***), and an ion corresponding either to lysophosphatidylethanolamine LPE(21:1) or isomeric lysophosphatidylcholine LPC(18:1), negative Spearman r=−0.23 (two-tailed test p-value=1.09e-34, ***). D: Correlation values between LD540 and lipid intensities across 2,840 single-cells, grouped by the lipid class. Tri-(TG) and di-glycerides (DG) are the most correlated which is in line with their reported function to compose the hepatic LDs core. Tukey box plots for each lipid class: center line: median; box limits: upper and lower quartiles; whiskers: 1.5x interquartile range. E: Enriched lipid ontology terms (LO) for lipids with an LD540-correlation >0.1 and <−0.1. Enriched LO terms from lipids with a high correlation with LD540 show enrichment of the neutral lipid metabolism, TGs, DGs, and lipid droplet biology, which is expected and therefore serves as a supportive argument for the capacity of SpaceM to detect and quantify biologically relevant molecules.
Hepatocytes were stimulated with oleic and palmitic fatty acids (FA) to model NAFLD-specific lipid metabolism23. The LD accumulation was quantified by the neutral lipid fluorescent dye LD54024. SpaceM provided profiles of 740 metabolites from 2,840 single dHepaRG cells. UMAP and unsupervised clustering of metabolic profiles revealed two distinct subpopulations characterized by statistically different amounts of LDs (Figure 2B, Supp. Fig. 8). Correlating LD540 and lipid intensities provided the following markers: triglyceride TG(48:0) (Spearman r=0.38***) for a smaller subpopulation, and isomeric lipids lysophosphatidylethanolamine LPE(21:1) or lysophosphatidylcholine LPC(18:1) (Spearman r=−0.23***) for a bigger subpopulation (Figure 2C). Further marker analysis showed neutral lipids, including triglycerides and diglycerides, to be the strongest markers confirmed by lipid ontology enrichment analysis25 (Figure 2E). This is expected as these lipids are known components of hepatic LDs26,27 (Figure 2D). Interestingly, the analysis of lipids anti-correlated with LD540 indicated an enrichment of glycerophospholipids and localization to endoplasmic reticulum (ER), a likely manifestation of the ER stress in hepatic steatosis28.
Characterisation of metabolic states of lipid stimulated hepatocytes in an inflammatory environment
The lipid composition in hepatocytes is tightly regulated under homeostatic conditions. However, an excess of free fatty acids, together with additional stressors, including inflammatory cytokines, promotes aberrant lipid storage, ultimately leading to NASH.
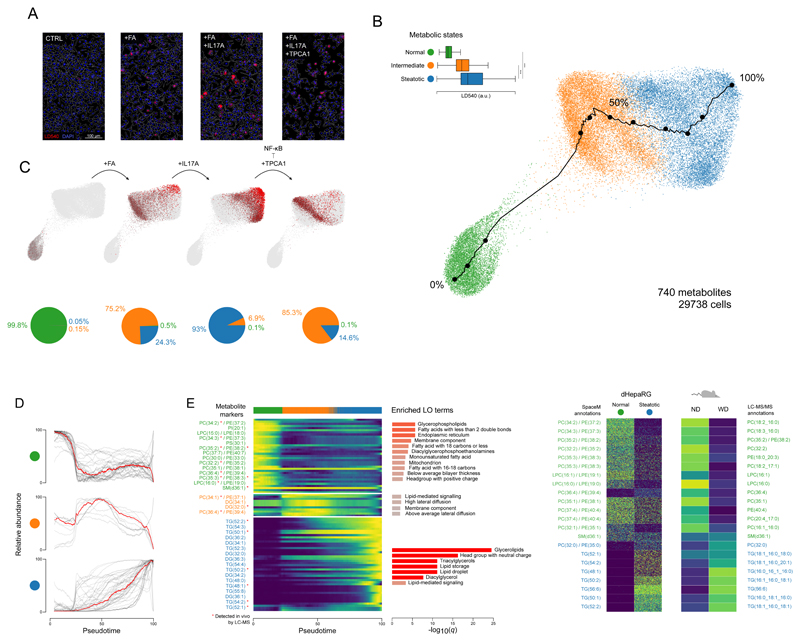
To investigate the metabolic remodeling of single hepatocytes during NAFLD to NASH transition, we considered four conditions (Figure 3A): (i) A control condition (CTRL) modelling the homeostatic state; (ii) stimulation with oleic and palmitic fatty acids (+FA) as in Figure 2, modelling non-inflammatory NAFLD; (iii) stimulation with FAs and inflammatory cytokine IL-17A (+FA+IL17A), modelling NASH and (iv) stimulation with FAs, IL-17A and TPCA-1 (+FA+IL17A+TPCA1), an inhibitor of NF-κB signalling, probing the metabolic response upon inhibition of IL-17A-induced inflammation.
Figure 3. SpaceM discovers and characterizes metabolic states of stimulated hepatocytes, cross-validated by preclinical NASH models.
A: Microscopy images of the human dHepaRG hepatocytes (CTRL), also stimulated with: fatty acids (+FA), further with IL-17A (+FA+IL17A), and further with an NF-kB inhibitor TPCA1 (+FA+IL17A+TPCA1); red: LD540 staining for lipid droplets; blue: Hoechst for nuclei; white: cells outlines. Representative images from 4 independent experiments per culture condition. B: PAGA visualization and unsupervised Leiden clustering of single-cell metabolic profiles (740 metabolites) reveal homeostatic, intermediate, and steatotic metabolic states of 29,738 hepatocytes. LD540 levels per metabolic states (homeostatic vs steatotic, two-tailed independent t-test p-value=0, ***; intermediate vs steatotic, two-tailed independent t-test p-value=1.4e-299, ***). Tukey box plots with center line: median; box limits: upper and lower quartiles; whiskers: 1.5x interquartile range. Black line shows a pseudotime trajectory from the homeostatic to steatotic states. C: Each condition separately, with cell values of LD540 in red, highlighting the gradient of the lipid droplet accumulation from the homeostatic to steatotic state. Pie charts show the proportions of the metabolic states in each condition. D: Normalized intensities of metabolic-state markers (t-statistic >60) for all cells along the pseudotime. E: Heatmap for the metabolic-state markers from C. Red star indicates validation by LC-MS/MS in the considered mouse model. For each state, the enriched lipid ontology (LO) terms are displayed. F: Semi-quantitative validation of selected metabolic markers of the homeostatic and steatotic states in an in vivo NASH mouse model with the normal diet (ND) vs Western diet (WD). For HepaRG, intensities of 2,500 randomly-selected cells are shown for each state. For the mouse model, average LC-MS intensity (n=3 replicates) per diet group is shown. Molecular names, left: putative MS1 annotations; right: structurally-validated by LC-MS/MS. ***denotes p-value<0.001.
SpaceM provided profiles of 740 metabolites from 29,738 hepatocytes with high reproducibility between replicates (Supp. Fig. 9). Unsupervised clustering of the metabolic profiles revealed three major clusters representing the homeostatic, intermediate and steatotic metabolic states (Figure 3B) with statistically different LD levels (Figure 3B). The CTRL condition was separated from FA-stimulated conditions and constitutes its own “homeostatic” metabolic state. Among +FA cells, 75.2% and 24.3% reside in the intermediate and steatotic states, respectively. Induction of inflammatory signalling with IL-17A drastically affected hepatocytes, with 93% of +FA+IL17A hepatocytes residing in the steatotic state. Of note, inhibition of NF-κB signalling by TPCA-1 prevents the uniform metabolic response seen upon stimulation with IL-17A as 75.2% of hepatocytes remain in the intermediate state (Figure 3C).
LD levels exhibit a higher variation in the +FA+IL17A condition (coefficient of variation, CV=0.54) compared to the +FA and +FA+IL17A+TPCA1 conditions (CV=0.37 and 0.45, respectively) (Supp. Fig. 10A, 10C). Conversely, individual lipids showed a higher variability of their single-cell intensities (showing a bimodal distribution) in the +FA and +FA+IL17A+TPCA1 conditions compared to the +FA+IL17A condition (showing a unimodal distribution) (Supp. Fig. 10B, 10C). Together, these results indicate that inducing inflammatory signalling in differentiated human hepatocytes with IL-17A increases cell-cell heterogeneity of LD levels but reduces heterogeneity of individual neutral lipids.
Pseudo-time analysis of single-cell metabolic profiles allowed us to characterize the progression of hepatocytes from the homeostatic to the steatotic state, (black line in Figure 3B). Metabolic state markers change gradually along pseudotime, indicating metabolic heterogeneity within every state (Figure 3D). Most markers of the homeostatic state are phospholipids such as phosphatidylcholines, phosphatidylethanolamines and phosphatidylinositols covering housekeeping metabolic functions including membrane composition of ER and mitochondria (Figure 3E). On the other hand, metabolic markers of the steatotic state are mostly neutral lipids such as triglycerides and diglycerides with a functional relevance in lipid storage, LD formation and lipid-mediated signalling.
The metabolic-state-markers were further validated using LC-MS/MS-based lipidomics of hepatic tissues from normal diet (ND) and Western diet (WD) mice. This helped perform MS/MS structural analysis and compare to the bulk levels of the markers in vivo (Figure 3F). The differences discovered by SpaceM were highly consistent with the marker levels in ND and WD mice. These data demonstrate that the metabolic states and markers discovered in vitro recapitulate the metabolic shift observed in vivo in early NASH development.
Discussion
The topic of metabolic heterogeneity has attracted a wealth of attention2. Bottom-up efforts on metabolic characterization of cell types29 and neighboring cells30 as well as top-down efforts on creating metabolic atlases for major cell types4,31 and finding metabolic subtypes in cancer cell lines32 reinforce this interest. However, these efforts lack single-cell measurements to complement either bulk metabolomics or metabolic modeling. We aim to contribute to bridging this gap and democratize single-cell metabolomics by providing SpaceM, a method for single-cell metabolomics of cultured cells. For each cell, SpaceM delivers a metabolic profile as well as morphometric properties and fluorescence intensities, can analyse thousands of cells each sampled in situ, is applicable to a broad range of adherent cell types cultured according to commonly-used practices and avoids any cell perturbation prior to cell fixation.
SpaceM complements other recently proposed single-cell metabolomics methods by delivering unique advantages in the context of analysis of cultured adherent cells. SpaceM offers higher throughput compared to cell microsampling followed by ultra-high-sensitive mass spectrometry33,34. Compared to laser-capture microdissection coupled with mass spectrometry e.g. through the liquid vortex capture or open port interface35,36, SpaceM focuses on the application to cultured cells (as compared to selected cells within a tissue section), offers a higher throughput, and delivers single-cell microscopy readouts.
Using imaging mass spectrometry is emerging as a viable option for single-cell metabolomics 9,12,13,37–39. Compared to one prominent example which uses a microarray for near-single-cell isolation 38, SpaceM allows cells to be analyzed in their native culturing environment thus avoiding any perturbation and delivering microscopy-based morphometric properties and cell-cell relationships. Compared to another prominent example, the MicroMS method using microscopy-guided cell ablation40,41, SpaceM offers several advantages. First, SpaceM can use any modern MALDI-imaging source without the need for a custom laser control. This is achieved by registering pre- and post-MALDI microscopy images with a reproducible subcellular precision in a fully automated way (Extended Data Fig. 2). Second, unlike MicroMS that requires cell dissociation, isolation, or reseeding to achieve sufficient spacing between cells11,40, SpaceM can directly analyze cells cultured at a high confluency in their native growth context by avoiding perturbations prior to fixation which are known to otherwise cause stress responses, metabolome alterations, and cell damages. This provides a substantial advantage for various commonly-used cell types such as used in our study (e.g. HeLa). Third, estimation of ablated cellular parts with a subcellular precision allowed us to develop a novel signal processing pipeline (Extended Data Fig 4) which provides a clear improvement of the co-cultured NIH3T3 and HeLa cell type classification, used here as a proxy for single-cell resolution (Supp. Fig. 6B). We further proposed a model for validating single-cell metabolomics of cell cultures by predicting co-cultured cell types, following the validation principles established in single-cell RNA-sequencing42. Addressing the current lack of established validation approaches, we hope this can benefit method development and validation in single-cell metabolomics.
Unbiased MALDI-imaging analysis -- as used in SpaceM -- leads to the cases of co-sampling neighboring cells by the same ablation mark. Although our normalization and ablation mark filtering were already designed to minimize the propagation of this effect to the intensities of the co-sampled cells, we have systematically investigated possible negative effects of the co-sampling. We have found that, with our normalization and filtration, the co-sampling only minimally affects the accuracy of the cell type prediction in spatially-heterogeneous co-cultures thus also indirectly confirming no negative effect on the single-cell resolution (Supp. Fig. 6). Further, we investigated the impact of the co-sampling on the metabolic states identified in hepatocytes (Supp. Fig. 11, data presented in Figure 2 and Figure 3 were computed with a sampling specificity value of 40%). Although excluding co-sampling ablation marks heavily reduced the number of cells (21.009 cells, compared 29.738 in Figure 3), it led to the identification of the same metabolic states (Supp. Fig. 11A). Moreover, it did not change the metabolic state markers although reducing their statistical significance, likely due the lower number of cells (Supp. Fig. 11A–B). Finally, excluding co-sampling ablation marks did not affect the balance of the metabolic states within each hepatocyte culture condition (Supp. Fig. 11C). Altogether, we conclude that the presented SpaceM processing approach delivers a substantial advantage over systematically excluding co-sampling ablation marks as it improves the single cell resolution while retaining more cells thus providing stronger statistical power.
Single-cell metabolomics of NAFLD/NASH was long-demanded to answer open questions about the associated liver metabolic reprogramming, motivated by the rising incidence of NAFLD/NASH, lack of treatment19 and high cell-cell heterogeneity of hepatic LDs18. Our results complement existing knowledge about the functional43, transcriptional44 and metabolic45 heterogeneity in hepatocytes during NAFLD-NASH progression- in which subpopulations emerge showing distinct functional43 and transcriptional44 phenotypes and plasticity of metabolic remodeling45.
We used a differentiated human hepatocyte cell line, dHepaRG which, unlike 2D in vitro cultured primary hepatocytes, exhibit a stable gene expression pattern, and is broadly used for investigating hepatic lipid metabolism and drug development16,17. The considered conditions model the stages of NAFLD-NASH progression: from homeostasis (condition CTRL) towards NAFLD (condition +FA), and further towards NASH (condition +FA+IL17A)19.
SpaceM helped characterize cell-cell metabolic heterogeneity of hepatocytes also within conditions. The clear separation of steatotic hepatocytes (Figure 2) from the rest of the population indicates a switch rather than a gradual change from the intermediate to the steatotic state. Previously, a steatotic subpopulation was associated with a protective function by accumulating LD-induced lipotoxicity that is advantageous for the whole population18.
Although lipid droplets can greatly vary in composition46, this heterogeneity cannot be resolved by lipophilic stainings as LD540. We explored the heterogeneity of levels of some neutral lipids in stimulated hepatocytes (Supp. Fig. 10). The intensities of selected steatotic markers showed a bimodal distribution not observed for LD540. This suggests that hepatocytes can be divided into two distinct populations, with low and high levels of steatotic markers (Supp. Fig. 10A–B). The bimodality of intermediate cells indicates that the overall bimodality cannot be explained by the presence of two states alone (Supp. Fig. 10B). This could either reflect a clustering artifact or biological heterogeneity of LD compositions for intermediate-state cells. Together, these results illustrate the importance of studying metabolism at single-cell resolution in combination with microscopy.
In line with previous reports, a number of steatotic markers were reproduced in our in vivo mouse model of NASH (Figure 3F). All but one validated triglyceride marker of the steatotic state are composed of palmitic acid (16:0), palmitoleic acid (16:1), and oleic acids (18:1) which were reported to increase in the liver of patients with NAFLD/NASH47. The steatotic-state-markers TG(50:1) and TG(48:1) were reported among the three most associated lipids with the fatty liver index (FLI, in the Fenland and NSHD studies) and with fat accumulation upon NASH48. Together, this demonstrates the in vivo relevance of our single-cell metabolomics analysis of dHepaRG hepatocytes.
SpaceM can differentiate cells within a state. The +FA steatotic cells (“benign” steatotic hepatocytes) and the +FA+IL17A steatotic cells (“inflammatory” steatotic hepatocytes) have little overlap in the PAGA plot (Figure 3B–C) suggesting a difference in their metabolic profiles. Differential analysis revealed that ceramide phosphocholines (sphingomyelins) were highly enriched in the “inflammatory” steatotic hepatocytes (Supp. Fig. 12). This is consistent with studies reporting an increase in sphingolipids including ceramides and sphingomyelins upon NASH compared to “benign” NAFLD in both mice and humans49,50. Although increased sphingolipids can be explained by the oxidative stress and systemic inflammation50, a mechanistic explanation is still lacking. Reproducing this effect in dHepaRG cells suggests the potential of in vitro single-cell metabolomics in deciphering the function of these bioactive lipids in NAFLD/NASH.
Overall, the open source SpaceM method not only fills a large yet empty niche in the booming field of single-cell metabolomics but already helped characterizing metabolic heterogeneity and states of stimulated dHepaRG hepatocytes, with the results validated in vivo and in line with previous reports. The simple design, low requirements, and compatibility with cell biology practices will likely pave a way towards democratizing single-cell metabolomics of cell cultures.
Methods
Co-culturing of HeLa and NIH3T3 cells
HeLa Kyoto H2B-mCherry and NIH3T3-GFP cells were cultured at 37 °C with 5% CO2, and were maintained in high glucose DMEM (1X Pen/Strep) (Gibco/ThermoFisher Scientific, Bremen, Germany) supplemented with 10% Fetal Bovine Serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco) and 1 mM sodium pyruvate (Gibco). Cells were trypsinized with 0.25% trypsin-EDTA (Gibco) and split 1:10 twice a week. Two technical replicates for the co-cultures and one replicate for monoculture were used. Trypsinized cells were counted and cells were seeded on 4-well-glass labtek chamber slides (Lab-Tek II, CC2) (ThermoFisher Scientific). In the co-culture experiment, an equal number of cells of each cell type was added into each well (4x105 cells/well). After 48h of incubation cells were washed with Phosphate Buffer Saline (PBS). After washing, the cells were fixed for 15 min with 4% paraformaldehyde (Sigma Aldrich, Darmstadt, Germany) at room temperature. Then the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (1μg/ml) (ThermoFisher Scientific) in PBS for 20 min at room temperature.
Hepatocytes culturing and stimulation
HepaRG cell culture and differentiation was performed as described earlier 51. Differentiated HepaRG (dHepaRG) cells were cultured on 4-well-glass chamber slides (Lab-Tek II, CC2, ThermoFisher Scientific, Bremen, Germany) (5.5x104 cells/well). The cells were stimulated with the fatty acids (opFAs): oleic acid (66 μM) and palmitic acid (33 μM), opFAs opFAs and the interleukin17-alpha (50 ng/ml) (Recombinant Human IL-17-alpha, RnD Systems), or opFAs and [5-(p-Fluorophenyl)-2-ureido]thiophene-3-carboxamide (TPCA1) (5nM) (Sigma Aldrich) in Williams E Medium (William’s Medium E, with stab. glutamine, without Phenol Red, with 2,24 g/l NaHCO3) (PAN Biotech) for 24 h. Cells grown in Williams E Medium without supplement for 24 h were considered as a negative control (CTRL). For each of those conditions, cells were seeded in three different wells which were considered as technical replicates (Table S1). After washing, cells were fixed for 15 min with 4% paraformaldehyde (Sigma Aldrich) at room temperature. Then the cells were washed and stained with Hoechst (1μg/ml) (Hoechst 33342) (ThermoFisher Scientific) and LD540 (0.1 μg/ml) 24 in PBS for 30 min at room temperature. After washing, cells were stored in dH2O at 4 °C for one night maximum.
Cell culture and LD540 staining of hepatocytes
Cell culture
2.2x105 HepaRG cells were cultured on 4-well-glass chamber slides (Lab-Tek II, CC2, Fisher Scientific) (5.5x104 cells/well) with or without fatty acids, oleic acid (66uM) and palmitic acid (33uM), IL-17A (final conc. 50ng/ml) (Recombinant HumanIL-17A, rnd Systems) and TPCA-1 (5nM) in Williams E Medium (William’s Medium E, with stab. Glutamine, without Phenol red, with 2,24 g/l NaHCO3, PAN Biotech) for 24h.
LD540 staining
After washing, cells were fixed for 15 min. with 4% paraformaldehyde at room temperature. Then the cells were washed and stained with Hoechst (1μg/ml) (Hoechst 33342, Thermo Fisher) and LD540 (0.1μg/ml) in PBS for 30 min. at room temperature. After washing, cells were imaged.
Microscopy acquisition
Cells were observed with the camera Nikon DS-Qi2 (Nikon Instruments) with the Plan Fluor 10x (NA 0.30) objective (Nikon Instruments) mounted on the Nikon Ti-E inverted microscope (Nikon Instruments) in bright-field and fluorescence (620 nm and 460 nm). The pixel size was 0.73 μm. The microscope was controlled using the Nikon NIS Elements software.
Mice and diets
4-6 weeks old male C57BL/6J mice were purchased from Charles River. Mice were housed at the at German Cancer Research Center (DKFZ) at a constant temperature of 20–24°C and 45–65% humidity with a 12-hour light/ 12-hour-dark cycle. 6-8 week-old male mice were fed ad libitum: normal diet (ND), western diet with trans-fat (WD-HTF) (Research Diets; D09100301 -40 kcal % fat (Primex shortening), 20 kcal % fructose, 2% cholesterol). Animals were maintained under specific-pathogen-free conditions and experiments were performed in accordance with German law and the governmental bodies, and with an approval from the Regierungspräsidium Karlsruhe ethical committee (G129/16, G7/17).
Cell image analysis
Cell segmentation and fluorescence quantification
Cells were segmented using a custom CellProfiler (v3.0.0) pipeline. Both morphological features and fluorescence intensities were quantified using CellProfiler and exported in .csv format for later interpretation.
Preparing cells for imaging
The plastic walls of the labtek were removed and the cells were dried in a Lab Companion™ Cabinet Vacuum Desiccator for 30min at room temperature and -0.08 MPa. After complete desiccation of the cells, pen marks are manually drawn on the glass slide using a black alcohol pen model 140s black (Edding, Ahrensburg, Germany) to keep track of the glass slide orientation and for image registration. The marks were drawn on the same side as the cells. Cells are kept at 4 °C until analysis. For the following experiments, the samples were analyzed by the microscopy and MALDI-imaging mass spectrometry following a randomized experimental design (Table S1).
Pre-MALDI bright-field and fluorescence microscopy of cells
Fixed cells were sequentially observed with the camera Nikon DS-Qi2 (Nikon Instruments) with the Plan Fluor 10x (NA 0.30) objective (Nikon Instruments) mounted on the Nikon Ti-E inverted microscope (Nikon Instruments) in bright-field and fluorescence (620 nm and 460 nm). The pixel size was 0.73 μm. The microscope was controlled using the Nikon NIS Elements software. The tiled acquisition of each cell culture area was performed using the JOB functionality of the NIS software. Stitching of tiled frames was performed using the FIJI stitching plugin 52.
MALDI imaging mass spectrometry
Relative humidity and temperature levels in the mass spectrometry room were monitored and controlled during the whole experiment and were within 44%-63% and 21.1-23.7 °C (Table S1). For the analysis of hepatocytes samples reported in Figure 2, Figure 3, Supp. Fig. 8, Supp. Fig. 9, Supp. Fig. 10, Supp. Fig. 11 and Supp. Fig. 12 (Conditions: Control, +FA, +FA+IL17a, +FA+IL17a+TPCA1), the 2,5-dihydroxybenzoic acid (DHB) matrix (Sigma Aldrich) 15mg/ml dissolved in 70% acetonitrile was applied onto the dried cells on the labtek slides by using a TM-Sprayer robotic sprayer (HTX Technologies, Carrboro, NC, USA). Spraying parameters were as following: temperature=100 °C, number of passes=8, flow rate=0.07 ml/min, velocity=1350 mm/min, track spacing=2 mm/min, pattern=CC, pressure=10 psi, gas flow rate=5 l/min, drying time=15 sec, nozzle height=41 mm. The estimated matrix density was 0.00311 mg/mm2. For the analysis of co-cultured cells in Figure 1, Supp. Fig. 4, Supp. Fig. 5, Supp. Fig. 6 and Supp. Fig. 7 the matrix 1,5-diaminonaphthalene (DAN) (Sigma Aldrich) 10mg/ml dissolved in 70% acetonitrile was applied onto the dried cells on the labtek slides by using the same TM-Sprayer robotic sprayer. Spraying parameters were as following: temperature=90 C°, number of passes=8, flow rate=0.07 ml/min, velocity=1350 mm/min, track spacing=3 mm/min, pattern=CC, pressure=10 psi, gas flow rate=2 l/min, drying time=15 sec, nozzle height=41 mm. The estimated matrix density was 0.001383 mg/mm2. For MALDI imaging mass spectrometry, the glass slides with the dried cells on them were mounted onto a custom slide adaptor and loaded into the AP-SMALDI source (Transmit, Giessen, Germany). The MALDI laser focus was optimized manually using the source cameras with the focused beam diameter estimated to be between 15.0 and 43.0 μm (mean equal to 29.9 μm, standard deviation equal to 8 μm). The x-y step size (distance between the centers of ablation marks) was set to 50 μM. For each pixel, the spectrum was accumulated from 30 laser shots at 60 Hz. Both the positive and negative mode MS analysis were performed using an QExactive Plus mass spectrometer (ThermoFisher Scientific) in the full scan mode in the mass range of 200-1100 m/z (resolving power R=140000 at m/z 200). MS parameters in the Tune software (version 2.5 Build 2042, ThermoFisher Scientific) were set to the spray voltage of 4.10 kV, S-Lens 80 eV, capillary temperature 250 C. The data was converted from the RAW format into the imzML format containing only centroided data using the ImageQuest software, v.1.1.0 (ThermoFisher Scientific). Metabolite annotation was performed using the METASPACE cloud software (https://metaspace2020.eu) implementing the bioinformatics methods for False Discovery Rate-controlled annotation published by us earlier 53 with the m/z tolerance of 3 ppm and FDR of 10%, 20%, and 50% against the HMDB metabolite database v2.5.
Post-MALDI microscopy to detect MALDI ablation marks
The cells were imaged in bright-field microscopy after MALDI-imaging using the same microscopy setup and parameters as described earlier in the pre-MALDI microscopy section to define the positions of the ablation marks with respect to the fiducial marks.
Association of laser ablation marks with single cells
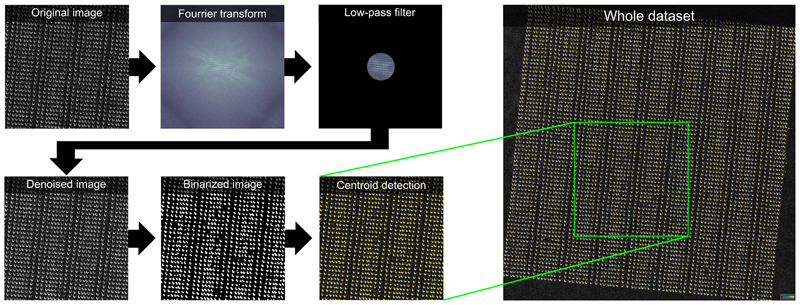
This step is the key part of the method as it solves the challenge that single cells are not visible in the post-MALDI microscopy images due to the opaque layer of MALDI matrix covering cells. Here, ablation marks left by the MALDI laser were associated with single cells in three steps: a) cells segmentation in the pre-MALDI microscopy images, b) detection of laser ablation marks in post-MALDI microscopy images, c) matching between ablation marks and MALDI mass spectra and d) co-registration of pre- and post-MALDI microscopy images to overlay the ablation marks with the segmented single cells.
In step a), cells were segmented using a custom pipeline in the CellProfiler software 54 where the DAPI staining channel was used to generate seeds for a region growing algorithm detecting cells boundaries in the LD540-staining channel. In step b), we first denoised the bright-field microscopy images by applying a low-pass filter in the 2D Fourier frequency domain, in particular to exploit both the regular distances between ablation marks as well as the repeated shape of the ablation mark itself. Then, we applied a contrast-enhancing filter (using the clip function from the Python module numpy) and Otsu’s thresholding method to binarize the image. Then, we applied morphological image analysis operations of closing and then opening to fill in the holes in the image and to remove individual noisy pixels. This provided estimations of the centers of mass of each ablation mark (Extended Data Fig 1). In step c), we fit a theoretical rectangular grid to the ablation marks. The numbers of X- and Y-grid steps were defined as set up during the MALDI acquisition. The center of the acquisition region was considered as the center of the grid. The orientation of the grid with respect to the post-MALDI microscopy image was optimized by finding an angle which resulted in the best overlap between the grid lines and the detected ablation marks. The X- and Y-spacing of the grid were optimized by minimizing the distance between the grid nodes and the center of mass of the nearest neighbor ablation mark. Then, only ablation marks which were the nearest neighbors to the grid nodes were taken and re-indexed (Extended Data Fig 3). This provided X- and Y-coordinates for each ablation mark associated with a collected MALDI spectrum. In order to improve estimations of the ablations marks areas used later for normalization, their segmentation was further improved by applying a custom region-growing algorithm implemented in Python. In step d), co-registration of pre- and post-MALDI microscopy images was done based on the pen marks drawn on the edge of the wells used as fiducials. We first segmented the pen marks in both pre- and post-MALDI bright-field microscopy images using Otsu’s intensity thresholding method. Then, we used the basin-hopping optimization algorithm (Python implementation from the scipy package v0.18.1) to find the best linear transformation matching the coordinates of the edges of the pen marks between the pre- and post-MALDI images (Extended Data Fig 2). The optimal linear transformation was applied to the post-MALDI microscopy images to map the ablation marks to the pre-MALDI microscopy images.
SpaceM processing
A normalized intensity of each metabolite in a single cell was constructed as follows. For each cell, we only considered the ablation marks which were overlapping with cells by at least 45% of their ablation area. This overlap is called sampling area. Additionally, the ablation marks that share less than 90% of their sampling area with the cell of interest are discarded. The metabolite intensities coming from an ablation mark were normalized by dividing them by the ratio of the sampling area to the area of the ablation mark. Finally, for each cell its normalized metabolite intensities were calculated as the weighted average normalized intensities of the associated ablation marks where the weights are defined as the ratio of the shared pixels (Extended Data Fig 4).
Selecting intracellular metabolites
We selected metabolite annotations corresponding to intracellular metabolites as follows. First, for each ablation mark we assigned to it the inside-cells label having values either of zero or one based on whether the mark has any overlap with any cell. Then, for each metabolite ion image, its intensities were binarized to zero-one values by selecting a threshold leading to the highest Spearman correlation with the inside-cells labels. The threshold value was found using the basin-hopping optimization algorithm. In order to consider only intracellular metabolites for further analysis, we selected those metabolite annotations whose binarized ion images were correlated with the inside-cells labels with the spearman correlation higher than 0.25. For the co-culture study we considered the metabolite annotations which were detected by METASPACE in at least 2 samples (out of 6 overall: 2 replicates of co-culture, 2 mono-cultures of NIH3T3 cells and 2 mono-cultures of HeLa cells) that led to 88 annotations. For each metabolite annotation, we pulled the ion images with the m/z tolerance of 3ppm from the imzML files.
Classification of ion images of METASPACE annotations into on- and off-sample classes
In the study of hepatocytes (Figure 2–3), we employed an advanced strategy to filter out background ions which might correspond either to matrix or contamination. First, we considered all ions annotated by METASPACE with 50% FDR. Then, two experts in MALDI-imaging performed manual tagging of these annotations (on-sample or off-sample) by loading every annotation in METASPACE and browsing the first 20 annotations from public METASPACE datasets. Based on the annotation ion images, the decision was made for each annotation whether it is more likely to be on- or off-sample. These taggings were then corrected using machine learning trained on the hepatocytes data following a molecular co-localization method we recently developed55. For every MALDI-imaging dataset, we have performed the following machine learning-based procedure to classify all its METASPACE annotations into on- and off-sample classes. First, for the ion image of each annotation we calculated its cosine co-localization with images of other annotations. As a result, every annotation was represented in a Euclidean space of the dimensions equal to the number of all considered annotations in this dataset. In this space, we have performed Support Vector Machine classification of on- and off-sample annotations by using the tags provided by experts as labels (using Python package scikit-learn 0.22.1). Overall 740 ions were classified as on-sample and were annotated by METASPACE.
Single cell data analysis
Single cell data from SpaceM were analyzed using Scanpy (v1.4.5)56. First cells with less than 5 metabolites were filtered out. The intensities were normalized per cell such that each cell has a total intensity equal to the median of total intensity per cell before normalization. The metabolite intensities are log10 transformed. Datasets from the same conditions are batch corrected. For the co-culture datasets, the batch balanced kNN integration (BBkNN) is performed 56. For the hepatocytes datasets, the scanpy implementation of the combat batch correction has been used condition-wise (Supp. Fig. 9). The Principal Component Analysis (PCA) is then computed followed by scaling the metabolite intensities to unit variance and zero mean. A neighborhood graph of observations is computed as described in the UMAP implementation of 56 with the parameters n_neighbors=25 and metric=‘cosine’, the rest is set as default.
Cell type classification for the co-culture study
The spatio-molecular matrices from HeLa and NIH3T3 cells were generated using the SpaceM processing with the following ablation marks filter parameters: sampling proportion 45%, upper area limit 95th percentile, sampling specificity 90%. The assignment of the cell type based on the constitutive fluorescence of the cells (mCherry for HeLa, GFP for NIH3T3) was done by finding a separating linear boundary between the two populations (Supp. Fig. 4A). The resulting cell types provided the ground truth labels. The cells were classified with accuracy of 91.3% based on their metabolic profiles (88 metabolite intensities) using the Linear Discriminant Analysis (LDA) (Supp. Fig. 4D). The classification accuracy has been estimated by performing a 10 times-repeated stratified 10-fold cross validation with different randomization in each repetition (100 repetitions in total).
Analysis of hepatocytes single-cell data
The spatio-molecular matrices from hepatocytes were generated using the SpaceM processing with the following ablation marks filter parameters: sampling proportion 45%, upper area limit 95th percentile, sampling specicity 40%. After computation of the neighbor graph, Leiden unsupervised clustering was performed to define metabolic states. UMAP visualization was computed to initialize the partition-based graph abstraction (PAGA) layout56. The Leiden clusters defined the groups for the PAGA algorithm implemented in Scanpy 1.4.5. The ForceAtlas2 layout has been used for computing the positions of each cell using the PAGA layout as coordinates for initialization (Supp. Fig. 9). To define the metabolite markers of a cluster, a t-test was performed between that cluster and the rest of the cells. Metabolites with a t-statistic superior to 60 were considered as markers. The pseudotime trajectory was computed using the coordinates of the PAGA graph, where the pseudotime value of a cell was equal to its normalized sorted index along the axis which shows the largest variation.
LC-MS/MS analysis on bulk cell cultures
Sample preparation
Lipids and fatty acids were extracted using the Folch method with chloroform:methanol (2.5:1). For lipidomics analysis, the dried samples were reconstituted in isopropanol:methanol (1:1) and injected 10uL into the LC-MS system. For metabolomics analysis, metabolites were extracted in 80% methanol and directly injected 20uL into the LC-MS system.
LC-MS/MS methods for lipidomics
LC-MS/MS analyses were performed on a Vanquish Ultra-High Performance Liquid Chromatography (UHPLC) system coupled to a Q-Exactive Plus High Resolution Mass Spectrometry (HRMS) (ThermoFisher Scientific) with an electrospray ionization (ESI) source operated in either positive or negative mode. The separation of lipids and fatty acids was carried out using an Agilent Poroshell EC-C18 column (3 x 50 mm; 2.7 μM) maintained at 40 °C at the flow rate of 0.26 ml/min. The mobile phase consisted of solvent A (acetonitrile−water (4:6)) and solvent B (isopropyl alcohol−acetonitrile (9:1)), which were buffered with either 10 mM ammonium acetate (for negative mode) or 10mM ammonium formate acidified with 0.1% formic acid (for positive mode). The UHPLC gradient was set at 20%, 20%, 45%, 52%, 66%, 70%, 75%, 97%, 97%, 20%, 20% of solvent B at the time points 0, 1.5, 4, 5, 7, 8, 10, 12, 15, 16, 19 min, respectively. Fatty acids and lipids were detected with the HRMS full scan at the mass resolving power R=35000 in the mass range of 100-1500 m/z. The data-dependent tandem (MS/MS) mass scans for five most intense ions (TOP5) were obtained along with full scans using higher energy collisional dissociation (HCD) with normalized collision energies of 20, 30 and 40 units at the mass resolving power R=17500. The MS parameters in the Tune software (ThermoFisher Scientific) were set as: spray voltage of 4 kV, sheath gas 30 and auxiliary gas 5 units, S-Lens 65 eV, capillary temperature 320 °C and vaporization temperature of auxiliary gas was 300 °C.
LC-MS/MS methods for metabolomics
LC-MS/MS metabolomics analysis was carried out using an Xbridge BEH Amide column (100X 2.1 mm; 2.5 μM) maintained at 40 °C at the flow rate of 0.3 ml/min. The mobile phase consisted of solvent A (7.5 mM ammonium acetate with 0.05% NH4OH) and solvent B (acetonitrile). The UHPLC gradient was set at 85%, 85%, 10%, 10%, 85%, 85% of solvent B at the time points 0, 2, 12, 14, 14.1, 6 min, respectively. Metabolites were detected with HRMS full scan at the mass resolving power R=70000 in mass range of 60-900 m/z. The data-dependent MS/MS mass scans were obtained along with full scans using HCD of normalized collision energies of 10, 20 and 30 units which were at the mass resolving power R=17500. The MS parameters in the Tune software (ThermoFisher Scientific) were set as: spray voltage of 4 kV (for negative mode 3.5 kV), sheath gas 30 and auxiliary gas 5 units, S-Lens 65 eV, capillary temperature 320 °C and vaporization temperature of auxiliary gas was 300 °C. Data was acquired in the full scan mode and MS/MS mass spectra for TOP5 precursor ions.
LC-MS/MS validation of METASPACE annotations
LC-MS/MS validation of lipid and metabolite METASPACE annotations was performed either by comparing retention times, exact m/z (MS) and fragmentation pattern (MS/MS) spectra with authentic standards or by matching MS/MS spectra with the EMBL Metabolomics Core Facility (MCF) spectral library (available at http://curatr.mcf.embl.de/) and public spectral libraries (LipidBlast, LIPID MAPS and mzCloud). The details of annotation validation are summarized in Supplementary Data S1. The structural annotation procedure for head groups (HD) and fatty acid side chains (SD) is described in detail in 53.
LC-MS/MS analysis of mouse liver tissue
Lipid extraction
Frozen tissue was first ground to a powder (to homogenize the tissue) using a hand-held mortar and pestle and the ground tissue was then weighed. Before Lipid Extraction, 10 uL of the Splash Lipidomix Mass Spec standard (Avanti Polar Lipids, Alabama, U.S.A) was added to the ground tissue. 500 ul of chloroform was then added followed by 20 s of vortexing. For phase separation, 200 uL of water was added to each sample followed by 20 s vortexing. Samples were then kept in the cold for 10 minutes followed by then added to induce phase separation. The mixture was then centrifuging for 5 minutes at 600 rpm. The lipid layer (bottom layer) was then pipetted out and the solvent dried down under a stream of Nitrogen. Prior to LC-MS analysis, the lipid extract was reconstituted using 50:50 isopropanol: methanol. All samples were normalized to a w/v of 1 mg/ml.
LC-MS/MS analysis
Lipid extracts were separated on a Kinetex C18 2.1 x 100 mm, 2.6 μm column (Phenomenex, Aschaffenburg, De). Separation was achieved by gradient elution on a binary solvent Vanquish UHPLC (Thermo Fisher Scientific, Bremen, DE). Mobile Phase A consisted of ACN: H2O (60:40) while mobile phase B consisted of IPA: ACN (90:10). For positive ionization, the mobile phases were modified with 10 mM ammonium formate and 0.1% formic acid while for the negative ionization mode, the mobile phases were modified with 5 mM ammonium acetate and 0.1% acetic acid. A flow rate of 260 μL/min was used for the separation and the column and sample tray were held constant at 30 °C and 4 °C respectively.
MS Instrumentation
MS analysis was performed on a Q-Exactive plus Mass Spectrometer (Thermo Fisher Scientific, Bremen, DE) equipped with a heated electrospray ionization probe. In both the positive and negative ionization modes, the S-Lens RF level was set to 65, the capillary temperature was set to 320 °C, the sheath gas flow was set to 30 units and the auxiliary gas was set to 5 units. The spray voltage was set to 3.5 kV in the negative ionization mode and 4.5 kV in the positive ionization mode. In both modes, full scan mass spectra (Scan Range m/z 100 -1500, R=35K) were acquired along with data-dependent (DDA) MS/MS spectra of the 10 most abundant ions. DDA MS/MS spectra were acquired using normalized collision energies of 30, 40, and 50 units (R= 17.5K and an isolation width = 1 m/z). The instrument was controlled using Xcalibur version 4.0.
Data analysis and initial lipid annotation
Progenesis Q1, version 2.3 (NonLinear Dynamics, a Waters Company, Newcastle upon Tyne, UK) was used for peak picking and for chromatographic alignment of all samples). Lipids were initially annotated from Progenesis Metascope and LipidBlast databases. Putative identification of lipids was done for ions that had MS/MS data.
Confirmation of lipid identification
A pooled sample was used for the lipid identification of statistically significant lipids. An inclusion list was created to acquire MS/MS spectra of the TGs lipids and spectra were acquired as described above. Full scan spectra of the pooled sample were also acquired by polarity switching between the positive and the negative ionization modes. Separate polarity switching experiments were acquired using mobile phases used in the positive ionization mode (containing ammonium formate and formic acid) analysis and those used for negative ionization mode (containing ammonium acetate and acetic acid analysis. For the polarity switching experiments, DDA MS/MS spectra were also collected in the negative mode. LC-MS/MS spectra of some of the significant lipids can be found in the Supplementary Data S1.
Statistics and Reproducibility
The reported p-values of the Pearson and Spearman correlations were calculated using the ‘scipy.stats.pearsonr’ and ‘scipy.stats.spearmanr’ functions from the Scipy 1.4.1 Python package. In both cases, the p-values were computed from a two-tailed test using the exact distribution of the correlation coefficient under the assumption that the values are drawn from independent normal distributions.
Data visualization
All plots were generated in Python, version 3.6.2, by using the packages matplotlib 3.1.3 and seaborn 0.8.1. The single cell analysis and visualizations were carried on using the Scanpy 1.4.5 module.
Extended Data
Extended Data Fig. 1.
Extended Data Fig. 2.
Extended Data Fig. 3.
Extended Data Fig. 4.
Supplementary Material
Acknowledgements
We thank Carina Beatrice Vibe for her support and feedback on the manuscript, Mohammed Shahraz, Mans Ekelhof, and Andrew Palmer for help with MALDI-imaging, Andreas Eisenbarth for software development, Nassos Typas for advising on biology and providing access to a microscope, Bashir El Debs and Joel Selkrig for training on microscopy and cell culturing, METASPACE software development team (all EMBL), Megan Stanifer and Steeve Boulant (DKFZ) for training on cell culturing, Angela Andersen (Life Science Editors) and Tracy O’Connor (Helmholtz Center Munich) for scientific editing, Samantha Seah and Christoph Merten (EMBL) for providing NIH3T3-GFP, Fabian Merkel and Christian Häring (EMBL) for providing HeLa Kyoto H2B-mCherry. We thank other members of the Thesis Advisory Committee of L.R.: Anne-Claude Gavin (EMBL) and Britta Brügger (Heidelberg University). This work was funded in part by the European Union’s Horizon 2020 program under the agreements 634402, 777222 (T.A.) and 667273 (M.H.), the DKFZ-MOST cooperation program (M.H., M.S.), Darwin Trust of Edinburgh (S.T.), SFB Transregio grants 179, 209, 1335 and I&I Helmholtz Zukunftsthema (all M.H.), the ERC Consolidator grants HepatoMetaboPath (M.H.) and METACELL (T.A.), and ERC Proof of Concept Faith (M.H.). We thank all the reviewers and the editor for detailed feedback that helped improve the manuscript.
Footnotes
Author Contributions
L.R. and T.A. conceived the method. L.R. developed the method. S.T. conceived and performed the co-culture experiments. K.O. performed the on-sample analysis for hepatocytes. R.G. and P.P. performed LC-MS/MS validation. M.S. and M.H conceived the hepatocytes study. M.S. cultured and prepared hepatocytes. L.R. and T.A. conceived and performed data analyses. L.R., M.S., M.H. and T.A. interpreted data. L.R., M.S., M.H. and T.A. wrote the paper. T.A. supervised and coordinated the work.
Competing Interests
L.R. and T.A. are the inventors on a patent application describing a spatial single-cell metabolomics method (application in the E.U. EP3610267A1, U.S. US20200057049A1, Canada CA3059818A1, Australia AU2018252185A1, World Intellectual Property Organization (Patent Cooperation Treaty) WO2018189365A1).
Data availability
All MALDI-imaging mass spectrometry data as well as metabolite and lipid annotations and images are publicly available through METASPACE (https://metaspace2020.eu/project/Rappez_2021_SpaceM). The MALDI-imaging mass spectrometry data and LC-MS/MS datasets are available in the MetaboLights repository under the accession number MTBLS78 (https://www.ebi.ac.uk/metabolights/MTBLS78). The microscopy data is available at the EBI BioStudies repository under the accession number S-BSST369 (https://www.ebi.ac.uk/biostudies/studies/S-BSST369).
Code availability
The SpaceM codebase is accessible as Supplementary Software and on GitHub (https://github.com/alexandrovteam/SpaceM). The spatio-molecular matrices as well as the code for their downstream processing, including generation of the main figures are available on Google Collaboratory (https://colab.research.google.com/drive/1CKdHDUkGIpAcBzrSfuCodMF_l2xbVAKT).
References
- 1.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, DeBerardinis RJ. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019;30:434–446. doi: 10.1016/j.cmet.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, et al. The landscape of cancer cell line metabolism. Nat Med. 2019;25:850–860. doi: 10.1038/s41591-019-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marioni JC, Arendt D. How Single-Cell Genomics Is Changing Evolutionary and Developmental Biology. Annu Rev Cell Dev Biol. 2017;33:537–553. doi: 10.1146/annurev-cellbio-100616-060818. [DOI] [PubMed] [Google Scholar]
- 6.Pelkmans L. Cell Biology. Using cell-to-cell variability--a new era in molecular biology. Science. 2012;336:425–426. doi: 10.1126/science.1222161. [DOI] [PubMed] [Google Scholar]
- 7.Lee M-CW, et al. Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc Natl Acad Sci U S A. 2014;111:E4726–35. doi: 10.1073/pnas.1404656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell AB, Trapnell C, Bloom JD. Extreme heterogeneity of influenza virus infection in single cells. Elife. 2018;7 doi: 10.7554/eLife.32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan KD, Fyrestam J, Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst. 2019;144:782–793. doi: 10.1039/c8an01581c. [DOI] [PubMed] [Google Scholar]
- 10.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Profiling metabolites and peptides in single cells. Nat Methods. 2011;8:S20–9. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibáñez AJ, et al. Mass spectrometry-based metabolomics of single yeast cells. Proc Natl Acad Sci U S A. 2013;110:8790–8794. doi: 10.1073/pnas.1209302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi M, Philip MC, Yang N, Sweedler JV. Single Cell Neurometabolomics. ACS Chem Neurosci. 2018;9:40–50. doi: 10.1021/acschemneuro.7b00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali A, et al. Single-cell metabolomics by mass spectrometry: Advances, challenges, and future applications. Trends Analyt Chem. 2019;120:115436 [Google Scholar]
- 14.Gilmore IS, Heiles S, Pieterse CL. Metabolic Imaging at the Single-Cell Scale: Recent Advances in Mass Spectrometry Imaging. Annu Rev Anal Chem. 2019;12:1–24. doi: 10.1146/annurev-anchem-061318-115516. [DOI] [PubMed] [Google Scholar]
- 15.Lombard-Banek C, et al. In Vivo Subcellular Mass Spectrometry Enables Proteo-Metabolomic Single-Cell Systems Biology in a Chordate Embryo Developing to a Normally Behaving Tadpole (X. laevis)*. Angew Chem Int Ed Engl. 2021 doi: 10.1002/anie.202100923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belloni L, et al. Targeting a phospho-STAT3-miRNAs pathway improves vesicular hepatic steatosis in an in vitro and in vivo model. Sci Rep. 2018;8:13638. doi: 10.1038/s41598-018-31835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanner N, et al. Regulation of Drug Metabolism by the Interplay of Inflammatory Signaling, Steatosis, and Xeno-Sensing Receptors in HepaRG Cells. Drug Metab Dispos. 2018;46:326–335. doi: 10.1124/dmd.117.078675. [DOI] [PubMed] [Google Scholar]
- 18.Herms A, et al. Cell-to-cell heterogeneity in lipid droplets suggests a mechanism to reduce lipotoxicity. Curr Biol. 2013;23:1489–1496. doi: 10.1016/j.cub.2013.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019;16:411–428. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 20.Malehmir M, et al. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat Med. 2019;25:641–655. doi: 10.1038/s41591-019-0379-5. [DOI] [PubMed] [Google Scholar]
- 21.Alexandrov T. Spatial Metabolomics and Imaging Mass Spectrometry in the Age of Artificial Intelligence. Annu Rev BioMed Data Sci. 2020;3:61–87. doi: 10.1146/annurev-biodatasci-011420-031537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson NH, Tuck M, Van de Plas R, Caprioli RM. Advanced Registration and Analysis of MALDI Imaging Mass Spectrometry Measurements through Autofluorescence Microscopy. Anal Chem. 2018;90:12395–12403. doi: 10.1021/acs.analchem.8b02884. [DOI] [PubMed] [Google Scholar]
- 23.Wolf MJ, et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 2014;26:549–564. doi: 10.1016/j.ccell.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Spandl J, White DJ, Peychl J, Thiele C. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 2009;10:1579–1584. doi: 10.1111/j.1600-0854.2009.00980.x. [DOI] [PubMed] [Google Scholar]
- 25.Molenaar MR, et al. LION/web: a web-based ontology enrichment tool for lipidomic data analysis. Gigascience. 2019;8 doi: 10.1093/gigascience/giz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ress C, Kaser S. Mechanisms of intrahepatic triglyceride accumulatioN. World J Gastroenterol. 2016;22:1664–1673. doi: 10.3748/wjg.v22.i4.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gluchowski NL, Becuwe M, Walther TC, Farese RV., Jr Lipid droplets and liver disease: from basic biology to clinical implications. Nat Rev Gastroenterol Hepatol. 2017;14:343–355. doi: 10.1038/nrgastro.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baiceanu A, Mesdom P, Lagouge M, Foufelle F. Endoplasmic reticulum proteostasis in hepatic steatosis. Nat Rev Endocrinol. 2016;12:710–722. doi: 10.1038/nrendo.2016.124. [DOI] [PubMed] [Google Scholar]
- 29.Lau AN, et al. Dissecting cell type-specific metabolism in pancreatic ductal adenocarcinoma. bioRxiv. 2020:2020.03.09.984237. doi: 10.1101/2020.03.09.984237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Colman MJ, et al. Interplay between metabolic identities in the intestinal crypt supports stem cell functioN. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- 31.Robinson JL, et al. An atlas of human metabolism. Sci Signal. 2020;13 doi: 10.1126/scisignal.aaz1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daemen A, et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci U S A. 2015;112:E4410-7. doi: 10.1073/pnas.1501605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillaume-Gentil O, et al. Tunable Single-Cell Extraction for Molecular Analyses. Cell. 2016;166:506–516. doi: 10.1016/j.cell.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 34.Liu R, Pan N, Zhu Y, Yang Z. T-Probe: An Integrated Microscale Device for Online In Situ Single Cell Analysis and Metabolic Profiling Using Mass Spectrometry. Anal Chem. 2018;90:11078–11085. doi: 10.1021/acs.analchem.8b02927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cahill JF, Kertesz V, Van Berkel GJ. Laser dissection sampling modes for direct mass spectral analysis. Rapid CommuN Mass Spectrom. 2016;30:611–619. doi: 10.1002/rcm.7477. [DOI] [PubMed] [Google Scholar]
- 36.Cahill JF, Riba J, Kertesz V. Rapid, Untargeted Chemical Profiling of Single Cells in Their Native Environment. Anal Chem. 2019;91:6118–6126. doi: 10.1021/acs.analchem.9b00680. [DOI] [PubMed] [Google Scholar]
- 37.Rubakhin SS, Lanni EJ, Sweedler JV. Progress toward single cell metabolomics. Curr Opin Biotechnol. 2013;24:95–104. doi: 10.1016/j.copbio.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342:1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Vertes A. Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity. Angew Chem Int Ed Engl. 2018;57:4466–4477. doi: 10.1002/anie.201709719. [DOI] [PubMed] [Google Scholar]
- 40.Comi TJ, Neumann EK, Do TD, Sweedler JV. microMS: A Python Platform for Image-Guided Mass Spectrometry Profiling. J Am Soc Mass Spectrom. 2017;28:1919–1928. doi: 10.1007/s13361-017-1704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann EK, Comi TJ, Rubakhin SS, Sweedler JV. Lipid Heterogeneity between Astrocytes and Neurons Revealed by Single-Cell MALDI-MS Combined with Immunocytochemical ClassificatioN. Angew Chem Int Ed Engl. 2019;58:5910–5914. doi: 10.1002/anie.201812892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mereu E, et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0469-4. [DOI] [PubMed] [Google Scholar]
- 43.Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aizarani N, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019 doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall Z, et al. Lipid zonation and phospholipid remodeling in nonalcoholic fatty liver disease. Hepatology. 2017;65:1165–1180. doi: 10.1002/hep.28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiam AR, Beller M. The why, when and how of lipid droplet diversity. J Cell Sci. 2017;130:315–324. doi: 10.1242/jcs.192021. [DOI] [PubMed] [Google Scholar]
- 47.Araya J, et al. Increase in long-chain polyunsaturated fatty acid n -6/n -3 ratio in relation to hepatic steatosis in patients with non-alcoholic fatty liver disease. CliN Sci. 2004;106:635–643. doi: 10.1042/CS20030326. [DOI] [PubMed] [Google Scholar]
- 48.Sanders FWB, et al. Hepatic steatosis risk is partly driven by increased de novo lipogenesis following carbohydrate consumptioN. Genome Biol. 2018;19:79. doi: 10.1186/s13059-018-1439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito K, et al. Characterization of hepatic lipid profiles in a mouse model with nonalcoholic steatohepatitis and subsequent fibrosis. Sci Rep. 2015;5:12466. doi: 10.1038/srep12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Apostolopoulou M, et al. Specific Hepatic Sphingolipids Relate to Insulin Resistance, Oxidative Stress, and Inflammation in Nonalcoholic Steatohepatitis. Diabetes Care. 2018;41:1235–1243. doi: 10.2337/dc17-1318. [DOI] [PubMed] [Google Scholar]
- 51.Gripon P, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmer A, et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2017;14:57–60. doi: 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
- 54.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ovchinnikova K, Kovalev V, Stuart L, Alexandrov T. OffsampleAI: artificial intelligence approach to recognize off-sample mass spectrometry images. BMC Bioinformatics. 2020;21:129. doi: 10.1186/s12859-020-3425-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MALDI-imaging mass spectrometry data as well as metabolite and lipid annotations and images are publicly available through METASPACE (https://metaspace2020.eu/project/Rappez_2021_SpaceM). The MALDI-imaging mass spectrometry data and LC-MS/MS datasets are available in the MetaboLights repository under the accession number MTBLS78 (https://www.ebi.ac.uk/metabolights/MTBLS78). The microscopy data is available at the EBI BioStudies repository under the accession number S-BSST369 (https://www.ebi.ac.uk/biostudies/studies/S-BSST369).
The SpaceM codebase is accessible as Supplementary Software and on GitHub (https://github.com/alexandrovteam/SpaceM). The spatio-molecular matrices as well as the code for their downstream processing, including generation of the main figures are available on Google Collaboratory (https://colab.research.google.com/drive/1CKdHDUkGIpAcBzrSfuCodMF_l2xbVAKT).