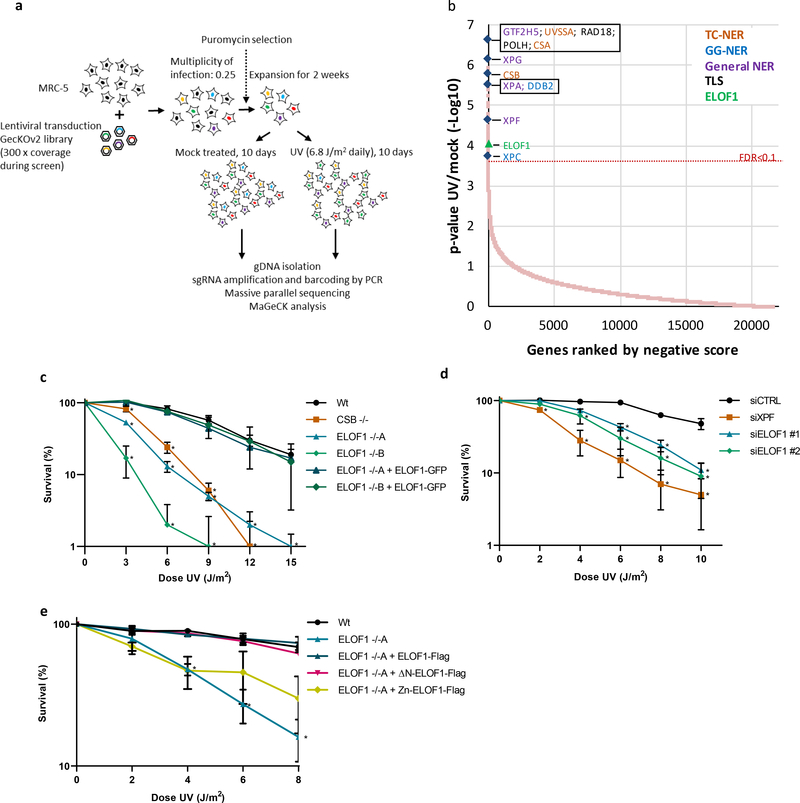
Figure 1. Genome-wide CRISPR/cas9 screen identifies ELOF1 as a factor involved in the UV-induced DNA damage response.
(a) Schematic of the CRISPR/cas9 screen. MRC-5 (SV40) cells were transduced with a lentiviral sgRNA library12. The resulting pool of gene-edited cells was split into a control and a UV irradiated group. Cells were respectively mock-treated or daily UV-irradiated with 6.8 J/m2 UV-C for 10 consecutive days, thereby maintaining ~50% cell confluency throughout the screen (Extended data Fig.1a). sgRNA abundance was determined by next-generation sequencing of PCR-amplified incorporated sgRNAs from the isolated genomic DNA of surviving cell pools47. UV-sensitive genes were identified by comparing the abundance in UV-irradiated cells over mock-treated cells using MAGeCK analysis. The screen was performed in duplicate. (b) UV-sensitive genes were ranked based on the gene-based P-value resulting from MaGecK analysis of the change in abundance of sgRNAs in UV-treated over mock-treated. Dotted line indicates FDR=0.1. Genes involved in NER or TLS are color-coded. (c) Relative colony survival of HCT116 wildtype (Wt) cells, indicated knock-out cells (−/−) or rescued cells exposed to the indicated doses of UV-C. (d) Relative colony survival of MRC-5 cells transfected with indicated siRNAs following exposure to the indicated doses of UV-C. (e) Relative colony survival of HCT116 ELOF1 KO cells with expression of the indicated ELOF1 mutants following exposure to the indicated doses of UV-C. Zn: zinc-finger mutant, ΔN: deletion of N-terminus. Data shown in c-e represent average ± SEM (n = 3 independent experiments) *P≤0.05 relative to Wt/siCTRL analyzed by one-sided unpaired T-test. Numerical data are provided in source data fig. 1.