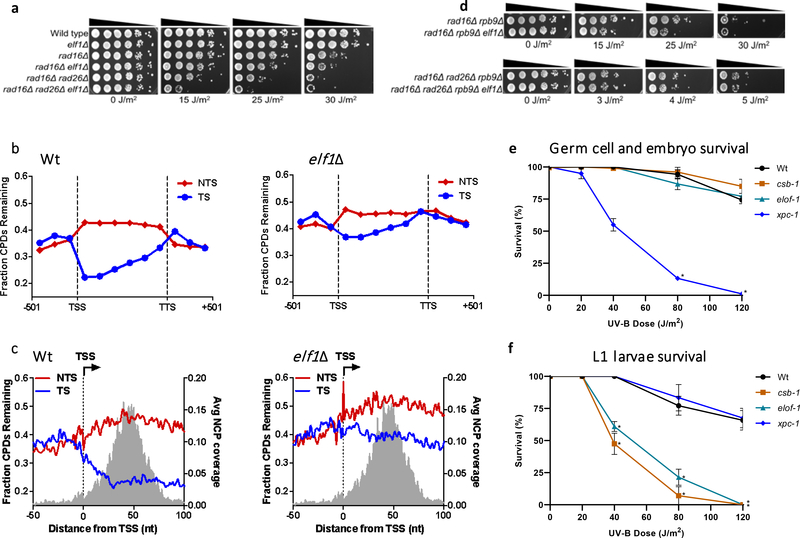
Figure 4. ELOF1 is an evolutionary-conserved TC-NER factor.
(a) Indicated mutant yeast strains were serially 10-fold diluted, spotted, and exposed to the indicated UV-C doses. Spot assay has been performed three times with similar results. (b) CPD-seq analysis of Wt (left) and elf1Δ mutant (right) yeast showing the average fraction of unrepaired CPDs remaining on the transcribed strand (TS) and non-transcribed strand (NTS) for ~5000 yeast genes following 2-hour repair relative to no repair. Each gene was divided in 6 equally-sized bins. Repair in flanking DNA upstream of the transcription start site (TSS) and downstream of the transcription termination site (TTS) is also depicted. (c) Close-up of CPD-seq repair data near the TSS in Wt (left) and elf1Δ mutant (right) cells. Nucleosome positioning data50 is shown for reference. CPD-seq has been executed once. (d) Indicated mutant yeast strains were serially 10-fold diluted, spotted, and exposed to the indicated UV-C doses. Spot assay has been performed three times with similar results. (e) C. elegans germ cell and embryo UV survival assay, measuring GG-NER activity, of wild type, csb-1, xpc-1, and elof-1 animals. The percentages of hatched eggs (survival) are plotted against the applied UV-B doses. (f) L1 larvae UV survival assay, measuring TC-NER activity, of wildtype, csb-1, xpc-1 and elof-1 animals. The percentages of animals that developed beyond the L2 stage (survival) are plotted against the applied UV-B doses. In e and f, the mean survival ± SEM of n = three independent experiments each performed in quintuple is depicted. *p≤0.05, ****p≤0.0001. Numerical data are provided in source data fig. 4.