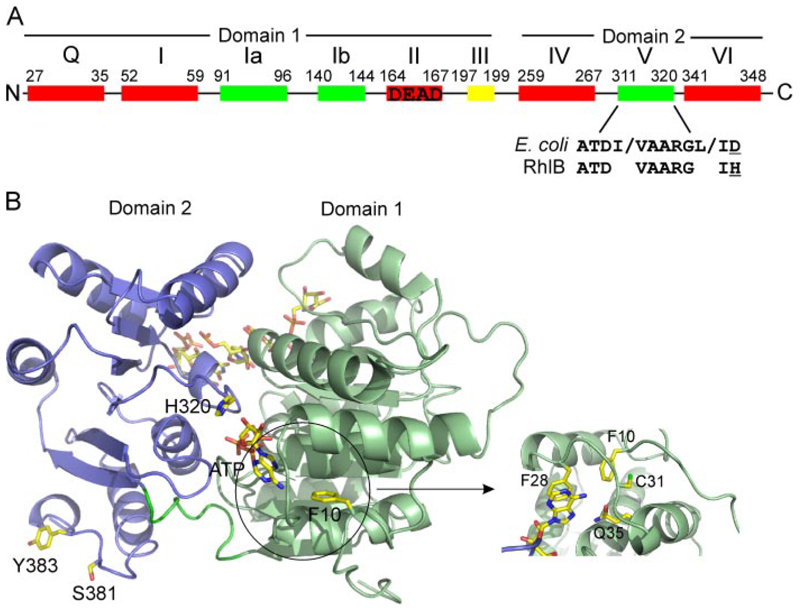
Figure 1.
A, location of the nine conserved sequence motifs in the DEAD box helicase RhlB. Color scheme indicates motifs involved in ATP binding and hydrolysis (red), RNA binding (green), and ATP-induced conformational change (yellow). Motif II is the Walker B motif, DEAD. The consensus amino acid sequence of motif V for E. coli DEAD box helicases is shown together with the sequence of RhlB highlighting the change in the final amino acid (underlined). B, homology model of RhlB, with domains 1 and 2 colored green and blue, respectively. The ATP, RNA, and locations of the residues mutated in this study are shown in stick representation. The inset is a close-up of the helix-loop-helix that forms theQ-motif in DEAD box helicases, with Phe-28 stacking with the adenine base and the invariantly conserved Gln-35 residue positioned to form hydrogen bonds with the N-6 and N-7 positions of the adenine base.