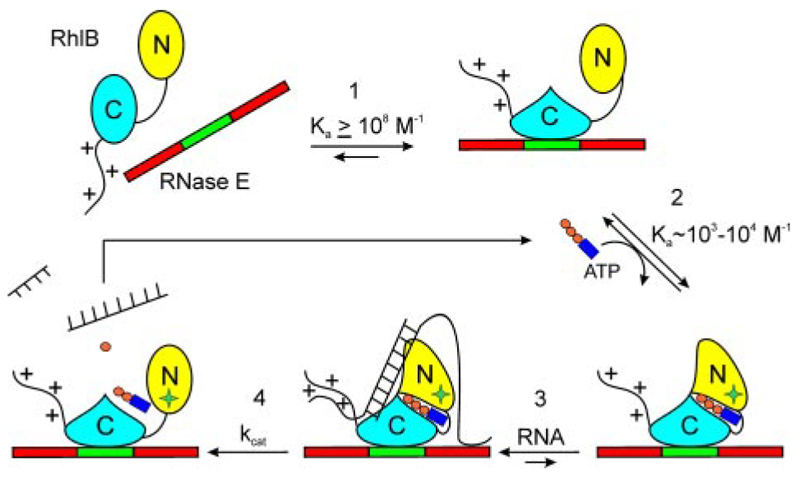
Figure 6. A schematic indicating a possible sequence of events in RhlB unwinding of duplex RNA.
1) RhlB binds to a surface of RNase E encompassing residues 696-762 (green) causing a possible change in structure of the C-terminal domain, which transmits to the ATP binding site.2) The binding of ATP induces association of the N-terminal and C-terminal domains; this perhaps causes a conformational switch in the surface of the N-terminal domain that formsan additional RNA binding site. The conformational switch is communicated through the Q-motif and Phe-10. This accounts for the inhibitory effect of RNA binding on ATP hydrolysis by the Phe-10 mutants. 3) The C-ter-minal tail of RhlB and the flanking RNA-bindings sites in RNase E (red) both interact with RNA. Deleting the tail eliminates ATPase activity (Table 3), showing that it plays a crucial role in the cycle. 4) Hydrolysis of ATP changes the surface, causing a change in the interaction with the RNA.