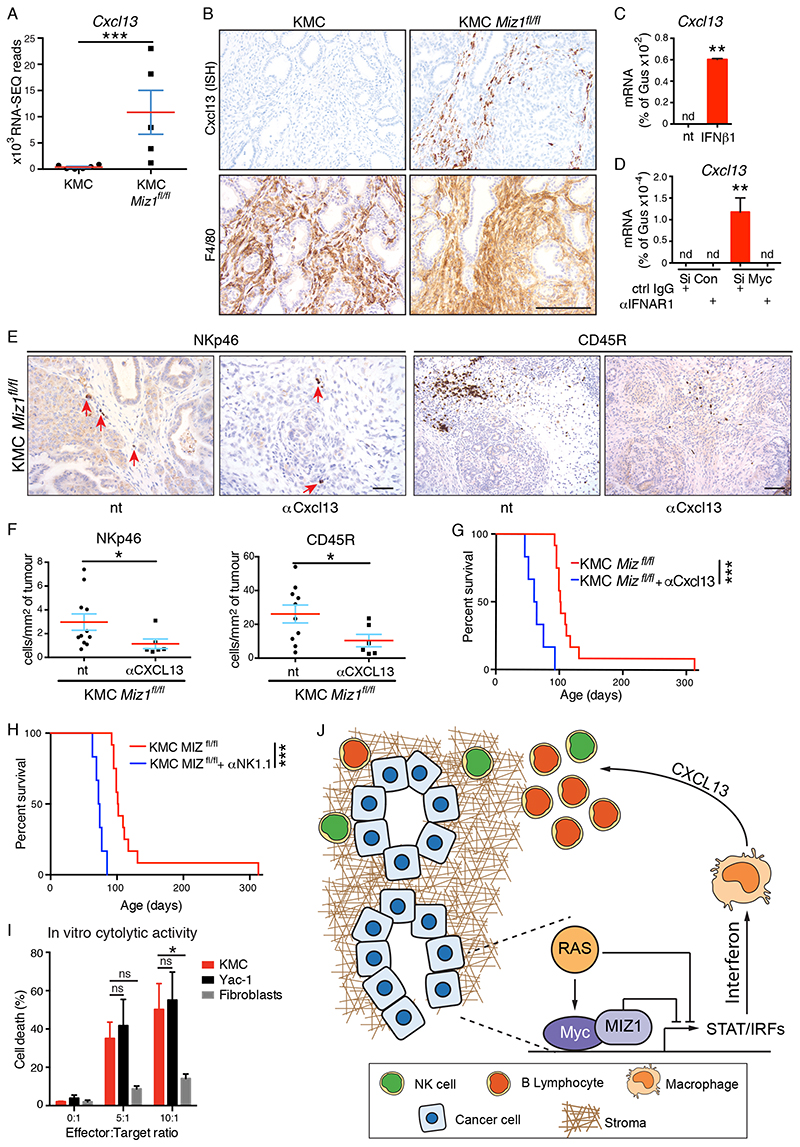
Figure 6. Cxcl13 mediates Interferon signalling to B and NK cells in PDAC.
A) Normalised RNA-SEQ reads of Cxcl13 expression in KMC (N=6) and KMC Miz1fl/fl (N=5) pancreatic tumours. B) In situ hybridization (ISH) analysis of Cxcl13-expressing cells in KMC and KMC-Miz1fl/fl tumours. Lower panels show IHC for F4/80. Scale bar = 100μm. C) RT-PCR analysis of Cxcl13 gene expression in bone marrow-derived macrophages upon treatment (10ng/ml for 4hrs) with recombinant mouse IFNβ1. Mean and SEM of technical replicates from 1 of 3 independent experiments. T test; nd = none detected. D) RT-PCR analysis of Cxcl13 gene expression in bone marrow-derived macrophages after 24hrs treatment with conditioned media from MYC-depleted or control KMC tumour cells. Macrophages were pre-treated (20μg/ml, overnight) with IFNAR1 blocking antibody or isotype control, where indicated. Mean and SEM of technical replicates from 1 of 3 independent experiments. ANOVA and Tukey post-hoc test; nd = none detected. E) IHC detection of tumour infiltrating NK and B cells after 3 weeks blockade of Cxcl13 in KMC-Miz1fl/fl PDAC. Scale bars = 100μm. F) Quantification of tumour infiltrating NK (NKp46+) and B (CD45R+) cells in untreated (N=11 & 10, respectively, from Figure 5G) and anti-Cxcl13-treated (N=6) KMC-Miz1fl/fl PDAC. Mann Whitney test. G) Overall survival of KMC-Miz1fl/fl mice treated with (N=6) or without (N=12, from Fig. 5F) anti-Cxcl13 blocking antibody for 3 weeks. Mantel Cox logrank test (G, H). H) Overall survival of KMC-Miz1fl/fl mice treated with (N=6) or without (N=12, from Fig. 5F) anti-NK1.1 depleting antibody for 4 weeks. I) FACS analysis of Zombie NIR™ labelling of target cells (KMC, Yac-1, Fibroblasts) co-cultured with IL-2 stimulated, NK cell enriched splenocytes for 4hrs. Mean and SEM of 3 independent experiments shown. 2-way ANOVA and Tukey’s multiple comparisons test. J) Model showing the mechanism of MYC/Miz1-dependent immune evasion in PDAC. For all panels, *** denotes p<0.001, **denotes p<0.01, * denotes p<0.05, ns = not significant.