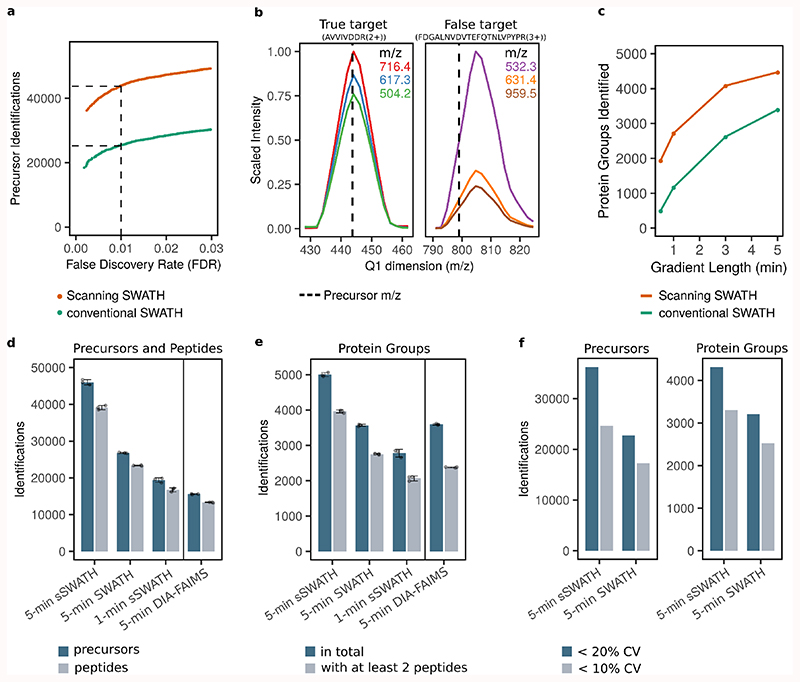
Figure 2. Scanning SWATH improves peptide identification in short gradients.
a. 10 µg human cell line digest was acquired with a Scanning SWATH method (10 m/z window) and a conventional stepped SWATH method 23 using a 5-minute high-flow (800 µL/min) LC gradient. The data were analyzed with DIA-NN using a two-species library, that contains human and Arabidopsis thaliana precursors, to validate the FDR experimentally (methods) 8,22. b. left panel: Q1 profiles of fragments corresponding to a true-positive target precursor (human) with a mass of 443.8 m/z (AVVIVDDR(2+)). b. right panel: Q1 profile of fragments corresponding to a false target (Arabidopsis thaliana precursor) with the precursor mass 799.1 m/z (FDGALNVDVTEFQTNLVPYPR(3+)). c. Number of protein groups identified (1% FDR) in a K562 cell lysate with Scanning SWATH and conventional stepped SWATH, using 5, 3, 1 minute and 30 second chromatographic gradients and adjusted duty cycles (Table S1, S2, S3). d. Number of precursors (peptides ionized to a specific charge) and peptides (stripped sequences) identified (1% FDR) in human cell lysates measured with different acquisition schemes and platforms. A K562 digest was analyzed with 5 and 1-minute gradient Scanning SWATH (“5-min sSWATH” and “1-min sSWATH”) and 5-minute conventional stepped SWATH (“5 min SWATH”). 10 µg was injected for the 5-minute gradients and 5 µg for the 1-minute gradient. To put the results into context, we compared them to a publicly available 5-minute gradient human cell line (HeLa) DIA dataset as recorded with an Evosep One system coupled to an Orbitrap Exploris 480 with FAIMS (“5-min DIA-FAIMS”) (PXD016662)25. Project-specific libraries and the same software settings were used for raw data analysis (Methods). Data are presented as mean +/- standard deviation (n=3 replicate injections). e. The number of protein groups (1% FDR) with at least one or two peptide identifications, respectively. Data are presented as mean +/- standard deviation (n=3 replicate injections). f. Number of precursors (left) and protein groups (right) quantified with a coefficient of variation (CV) below 20% and below 10% in triplicate injections. CV values were calculated from n = 3 replicate injections.