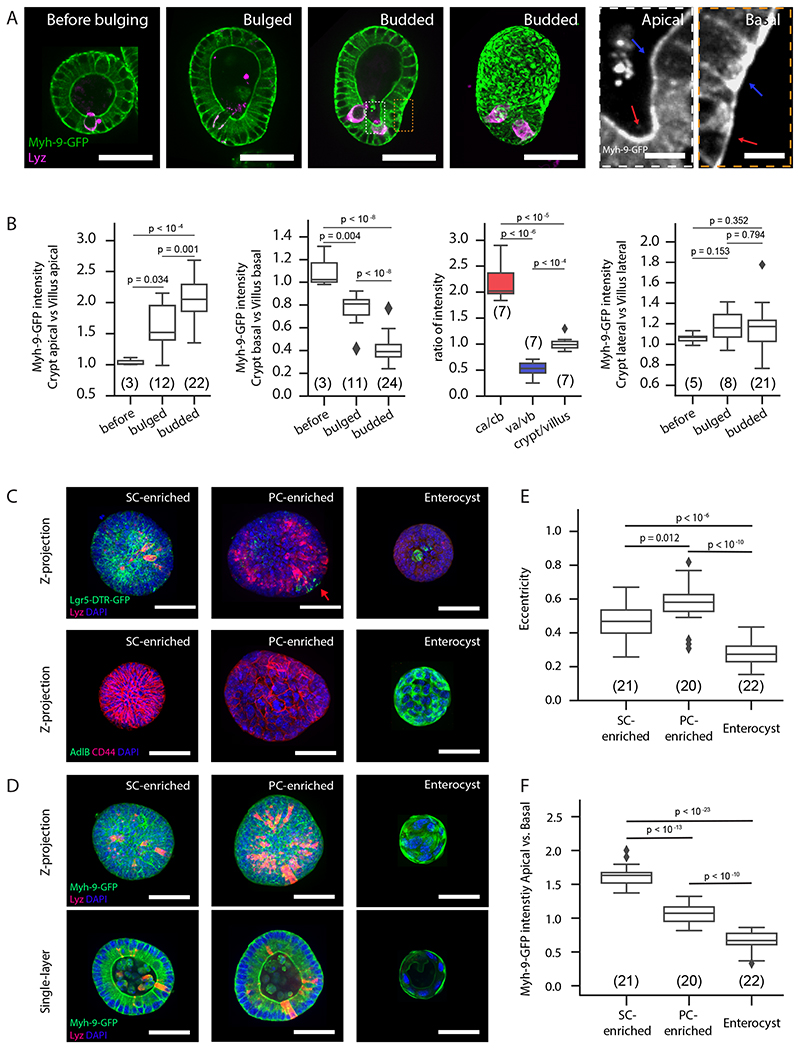
Figure 3. Myosin patterns in stem cells and enterocytes determine region-specific spontaneous curvature.
A. Myh-9-GFP in organoid before bulging, bulged, and budded organoids in the middle section, maximum z-projection in budded organoid, and zoom-in areas from dashed boxes. B. Plots for ratios of Myh-9-GFP intensity in cells. Abbreviations: ca, crypt apical; cb, crypt basal; va, villus apical; vb, villus basal. Arrows: villus (blue), crypt (red). C-F. Stem cell fate is responsible for apically enriched Myh-9 in crypt. C-D. Representative images for organoids treated by CHIR and VPA and enriched with stem cells (SC-enriched), treated by CHIR and DAPT and enriched with Paneth cells (PC-enriched) or enriched with enterocyte (enterocyst). C. Maximum z-projection of Lgr-5-DTR-GFP organoids with Lyz and DAPI staining (upper panels) and wild-type organoids with CD44, AdlB and DAPI staining (lower panels). D. Maximum z-projection (upper panels) and single-layer (lower panels) images of Myh-9-GFP organoids with Lyz and DAPI staining. E. Quantification of eccentricity from Maximum z-projection images in D. F. Quantification of Myh-9-GFP intensity from single-layer images in D. For box plots B, E, F n numbers are stated in brackets and represent organoids selected for multiple single-cell (B) or whole organoid (D) measurements. P-values (B, E and F) are determined by two-tailed t-test. Box-plot elements show 25% (Q1, upper bounds), 50% (median, black lines within the boxes) and 75% (Q3, lower bounds) quartiles, and whiskers denote 1.5× the interquartile range (maxima: Q3 + 1.5× (Q3-Q1); minima: Q1-1.5× (Q3-Q1)) with outliers (rhombuses). Experiments in A and C-E were repeated >3 times independently with similar results. Scale bars: A (organoid, 50μm; zoom-in, 5 μm), C and D (50 um).