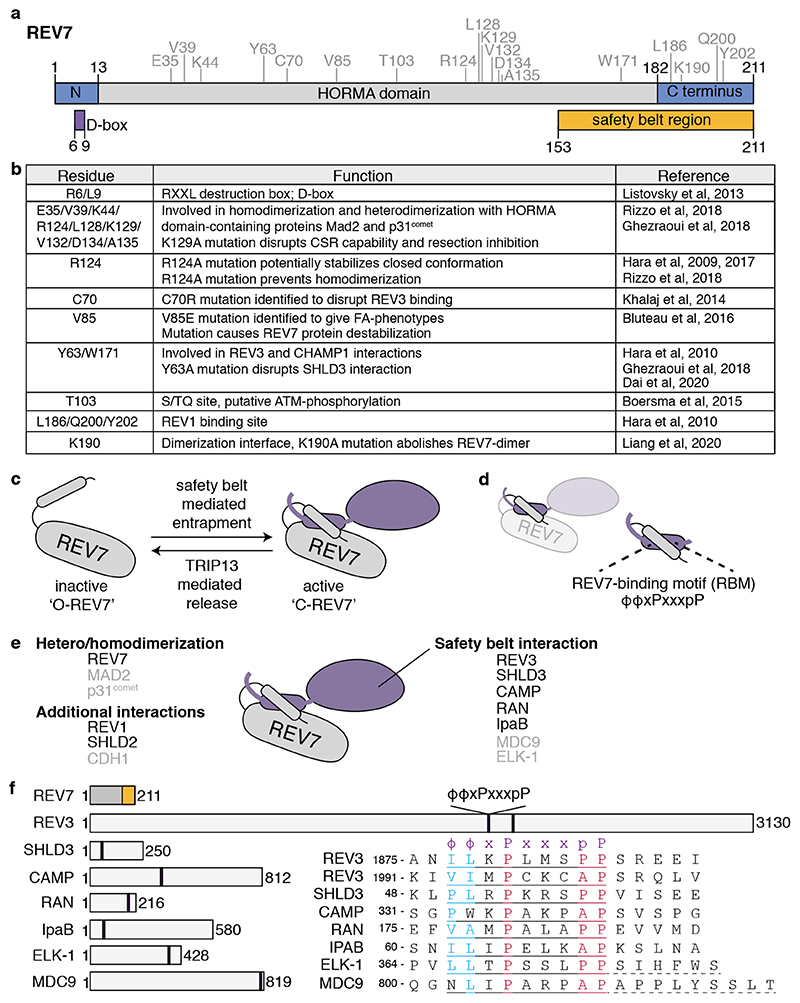
Figure 1. REV7 is a HORMA domain containing protein.
a. REV7 domain structure. Important structural features are indicated below, such as the destruction box (D-box) and the safety belt region (a.k.a. seatbelt). REV7 amino acid residues that have been described to be important for REV7 structure and function are indicated above in grey and are shown in (b). b. Table showing the different residues important for REV7 function. c. Schematic representation on how REV7 entraps peptides through its safety belt. Upon entrapment, REV7 undergoes a conformational switch from inactive, open-REV7 (‘O-REV7’) to active closed-REV7 (‘C-REV7’). The reverse reaction is mediated by the action of TRIP13, which thereby facilitates partner release. d. The REV7 safety belt entraps a REV7 Binding Motif (RBM) on its partner. e. Schematic representation of the different interaction-surfaces used by REV7 to interact with other proteins, also indicating what proteins have already been identified to interact with REV7 through each domain. REV7 dimerization appears essential for the interaction with SHLD2, and thereby could provide REV7 with an extra interaction-surface. For proteins indicated in black, the crystal structure in complex with REV7 has been described. f. Schematic representation of REV7 and proteins for which an RBM has been described, with their sizes in amino acids. The amino acids of the respective RBMs and their relative position within the protein are indicated. The RBM is defined as ϕϕxPxxxpP, in which ϕ denotes an aliphatic residue[15]. For ELK-1 and MDC9 additional residues are needed for the interaction with REV7, as indicated with dashed underline[15].