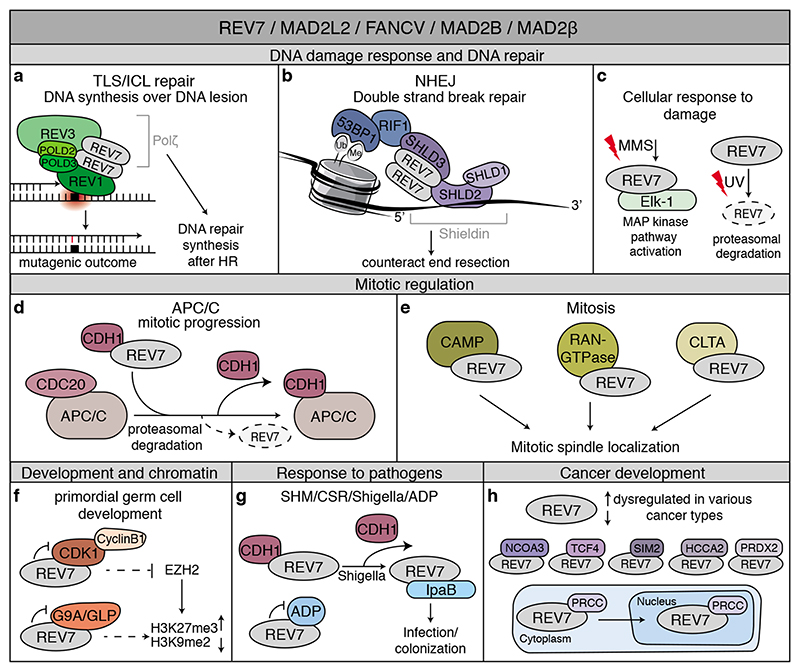
Figure 3. REV7: Jack of many trades.
Overview of the wide range of processes in which REV7 is involved through interactions with various proteins. See text, Table 1 and box 1 for details. a. REV7 functions in TLS by forming Polζ together with REV3, POLD2 and POLD3. Polζ also contributes to DNA repair synthesis after HR. b. The shieldin complex, consisting of REV7, SHLD3, SHLD2 and SHLD1, counteracts resection at DNA DSBs and thereby contributes to NHEJ. c. REV7 contributes to the cellular response to DNA damage. Upon DNA damage exposure by methyl methanesulfonate (MMS), REV7 contributes to MAP kinase pathway activation through the transcription factor Elk-1. Additionally, UV-damage induces degradation of REV7. d. REV7 binds to and sequesters the APC/C-coactivator CDH1, and thereby contributes to the correct timing of mitotic progression. e. REV7 interacts with CAMP, RAN GTPase and CLTA that colocalize with REV7 at the mitotic spindle. f. REV7 contributes to primordial germ cell development and chromatin regulation. In this context, REV7 was found to interact with G9A/GLP methyltransferases and Cdk1. g. REV7 contributes to somatic hypermutation (SHM) of immunoglobulin genes and immunoglobulin class-switch recombination (CSR) through functioning in TLS and shieldin. Additionally, REV7 interacts with IpaB and ADP. Together, REV7 thereby contributes to the cellular response to pathogens. h. REV7 is dysregulated in various cancer types. In addition, REV7 interacts with NCOA3, TCF4, SIM2, HCCA2 and PRDX2 in various cellular contexts or related to cancer development and treatment. The interaction of REV7 with PRCC contributes to REV7 subcellular localization.