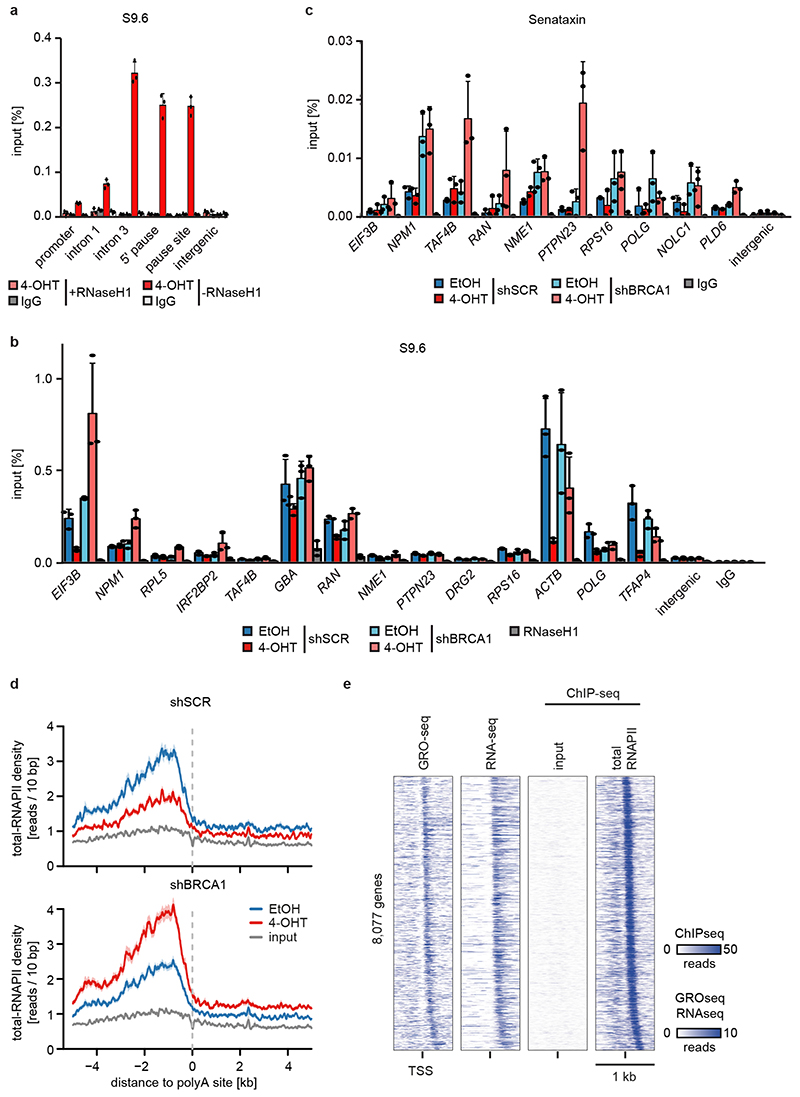
Extended Data Figure 8.
a: DNA-RNA-Immunoprecipitation (DRIP) using the S9.6 antibody, indicating R-loops at known loci within the ACTB gene. Digestion with RNaseH1 was used as control for specificity of the antibody and IgG as control for unspecific chromatin binding. Shown is the mean and error bars indicate SD of technical triplicates of one representative experiment (n=2).
b: DRIP documenting binding of the S9.6 R-loop antibody to the indicated loci upon depletion of BRCA1 and activation of MYCN for 4 h. Shown is the mean and error bars indicate SD of technical triplicates of one representative experiment (n=4). The panel shows the non-normalized data of Figure 3d.
c: ChIP of Senataxin at the indicated loci in SH-EP MYCNER cells after 4-OHT treatment (3 h) in control or BRCA1-depleted cells. Shown is the mean and error bars indicate SD of technical triplicates of one representative experiment (n=2 with two different antibodies).
d: RNAPII density (mean+SEM as shadow) around the first downstream polyA site in SH-EP MYCNER cells after BRCA1-depletion with shBRCA1#2 and MYCN-activation for 3h: 1,713 genes are shown. One representative experiment is shown (n=3).
e: Heat map of 8,077 TSSs of genes with an RNAPII-peak downstream of the start site. Reads originate from GRO-seq, mRNA-seq and total RNAPII-ChIP-seq samples and genes are sorted based on the distance from the TSS to the RNAPII-peak.