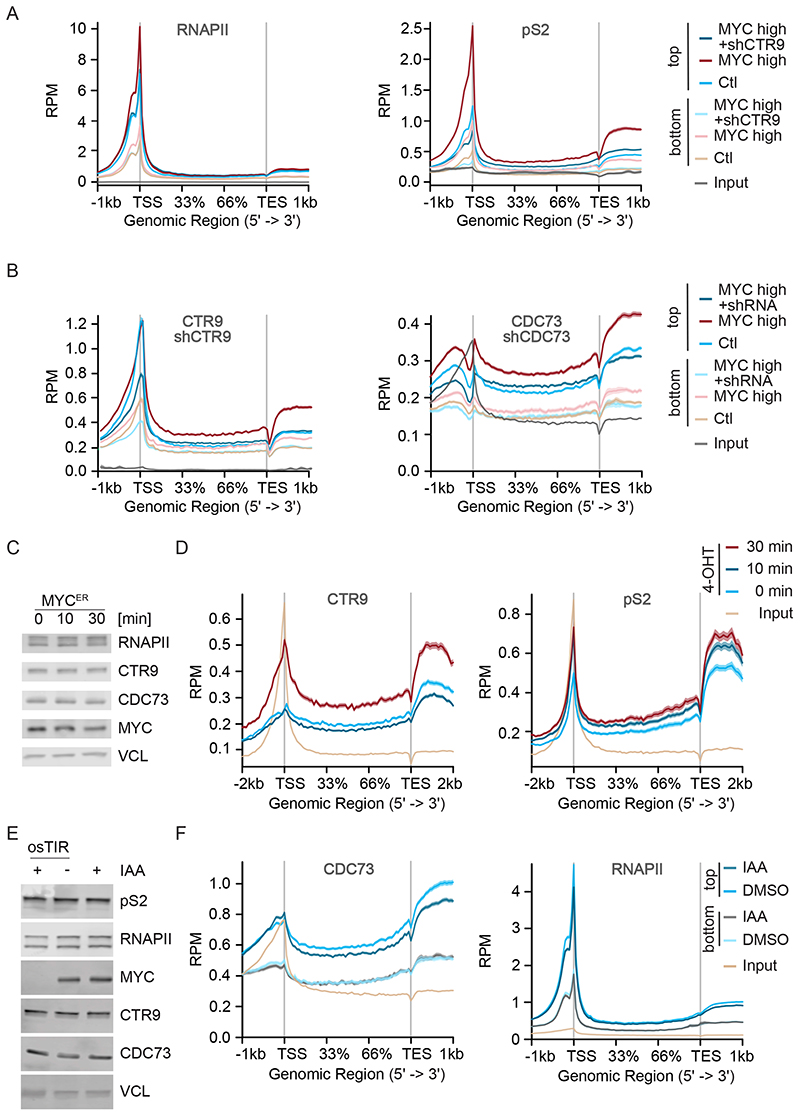
Figure 2. PAF1c is rapidly transferred from MYC on RNAPII.
A. Metagene plot of total RNAPII or pS2 RNAPII in a ChIP experiment in control U2OS cells or in cells stably expressing MYC and Dox inducible shCTR9; metagene plots of 7,479 most strongly MYC-bound (“top”) or 5,768 weakly MYC-bound (“bottom”) genes are shown (see STAR methods). Input shows 17,697 genes. Chromatin of six independent experiments was pooled for ChIP-sequencing.
B. Metagene plot of CTR9 or CDC73 binding to chromatin in a ChIP experiment, in control U2OS cells or in cells stably expressing MYC and Dox inducible shCTR9 or shCDC73; metagene plots of top or bottom MYC-bound genes. Input shows 17,697 genes. Chromatin of six independent experiments was pooled for ChIP-sequencing.
C. Immunoblot of U2OS cells expressing a MYCER chimeric protein with or without MYC activation upon addition of 4-OHT (200 nM) for 10 min or 30 min.
D. Metagene plot of CTR9 and pS2 in a ChIP-Rx experiment in U2OS MYCER cells treated as described in (C). The plot shows profiles of the top 4000 MYC bound genes (n=2).
E. Immunoblot of K562-MYC-AID erythroleukemia cells (n=2). Indole-3-acetic acid (IAA: 100 μM) was added for 30 min. Vinculin (VCL) was used as loading control (n=3).
F. Metagene plot of CDC73 or total RNAPII in a ChIP-Rx experiment in K562-MYC-AID cells treated as in (E). Metagene plots of top or bottom MYC-bound genes (n=2). Input shows 17,697 genes. See also Figure S2.