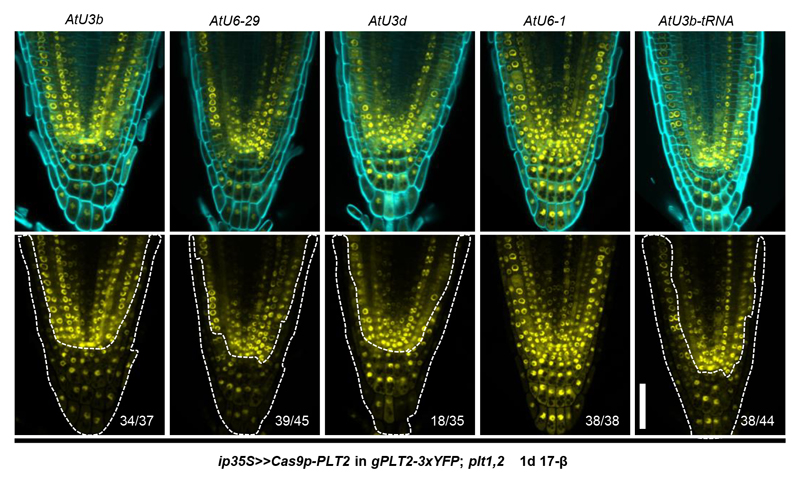
Extended Data Figure 4. sgRNA promoter identity affects editing efficiency in Arabidopsis roots.
For each construct, the indicated sgRNA promoter was used to drive transcription of sgRNA1, while ip35S was used to guide Cas9p transcription. AtU3b and AtU6-29 showed the highest editing efficiency in T1 seedlings after one-day of induction (1d 17-β). This may explain the preferred detection of deletion between target1 (AtU3b) and target4 (AtU6-29) when four sgRNAs were used in a single construct (Supplementary Fig. 4 and Extended Data Fig. 3a). Transcription of tRNA together with sgRNA1 under the AtU3b promoter also resulted in efficient PLT2 editing. White dotted lines mark the region with reduced YFP signal. This corresponds to the region where ip35S is active (Fig. 2a and Supplementary Fig. 3). Cell walls are highlighted by calcofluor. Numbers indicate the frequency of similar results in the independent T1 samples analyzed. All experiments were repeated three times. Scale bar, 50 μm.