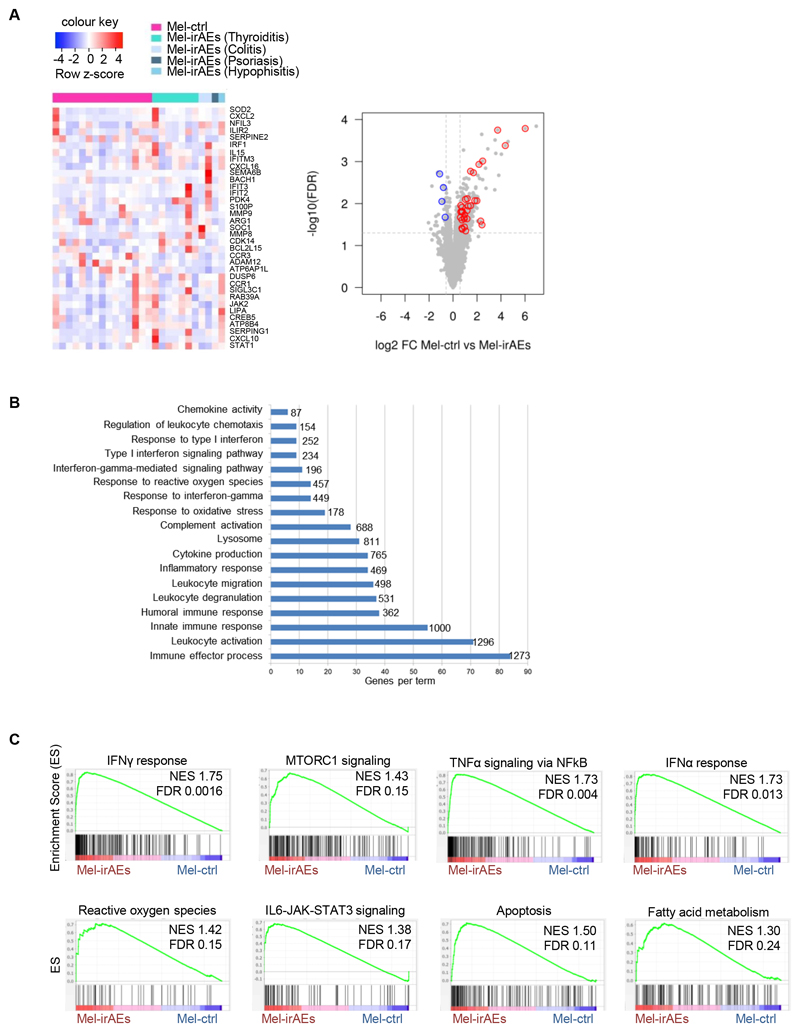
Figure 1. Treg inflammatory transcriptomic reprogramming in patients with advanced melanoma who develop irAEs.
Transcriptomic analysis of CD4+CD25+CD127– Tregs isolated from blood of advanced melanoma patients undergoing anti-PD1 treatment. (A) Heatmap with scaled cpm expression values (row z-score) (left) and volcano plot (right) of selected DEGs between ‘Mel-ctrl (n=15) vs Mel-irAEs (n=11)’. Categories of irAEs are listed on the heatmap. (B) Bar plot representing pathway analysis of DEGs from ‘Mel-ctrl vs Mel-irAEs’with adjusted p value ≤ 0.05. Each term size is listed on the end of each bar.(C) GSEA plot showing the enrichment of “IFNγ response” (NES 1.75, FDR 0.0016), “MTORC1 signaling” (NES 1.43, FDR 0.15). “Reactive oxygen species” (NES 1.42, FDR 0.15), “IL6-JAK-STAT3 signaling” (NES 1.38, FDR 0.17), “TNFα signaling via NFkB” (NES 1.73, FDR 0.004), “IFNα response” (NES 1.73, FDR 0.013), “Apoptosis” (NES 1.50, FDR 0.11) and “Fatty acid metabolism” (NES 1.30, FDR 0.24) gene set. For DEG analysis, the GLM statistical approach and the thresholds: |FC|≥1.5 and p<0.05 were used.