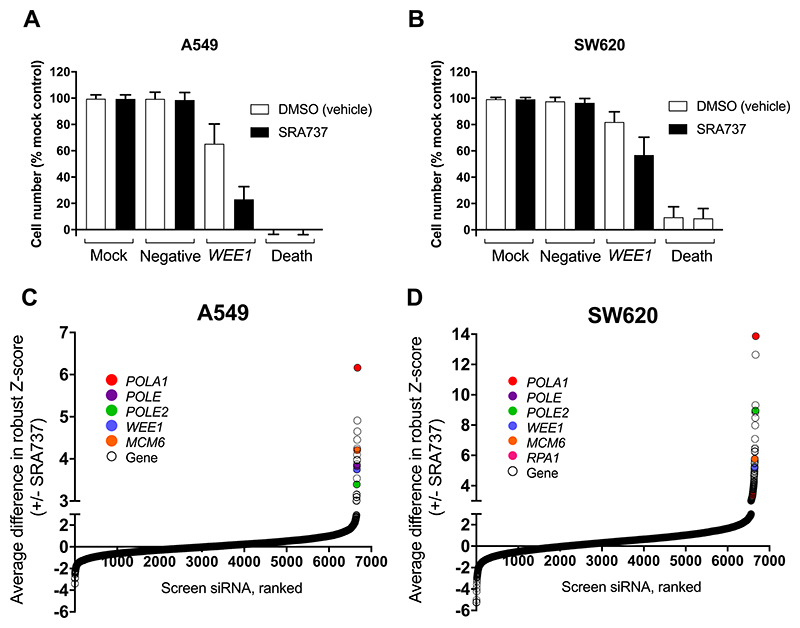
Figure 1. An siRNA screen identifies knockdown of DNA replication genes as synthetically lethal with CHK1 inhibition in human A549 NSCLC and SW620 colorectal cancer cell lines.
A and B Negative and positive control data for the siRNA library screen in human NSCLC A549 cells (A) and human colorectal cancer SW620 cells (B). Cell number is shown as a percentage of the mock control. Additional controls: Allstars negative siRNA (negative control for off target effects), WEE1 siRNA (positive control for synthetic lethality with SRA737) and death siRNA (positive control for transfection efficiency). C and D The average difference in robust Z-score for A549 (C) and SW620 (D) cells transfected with siRNA and treated with either DMSO (vehicle control) or SRA737. Top hits common to both cell lines are shown as closed coloured circles. A-D Library siRNA (25nm) were introduced into cells by reverse transfection. Mock cells were transfected with lipid only. Cells were then incubated for 48 h followed by treatment with either vehicle (DMSO) control or SRA737 (0.4 or 0.8 μM SRA737, A549 and SW620 respectively) for a further 84 h and then an SRB assay performed. Each cell line was screened with the siRNA library three times (3 biological repeats) to generate the data shown.