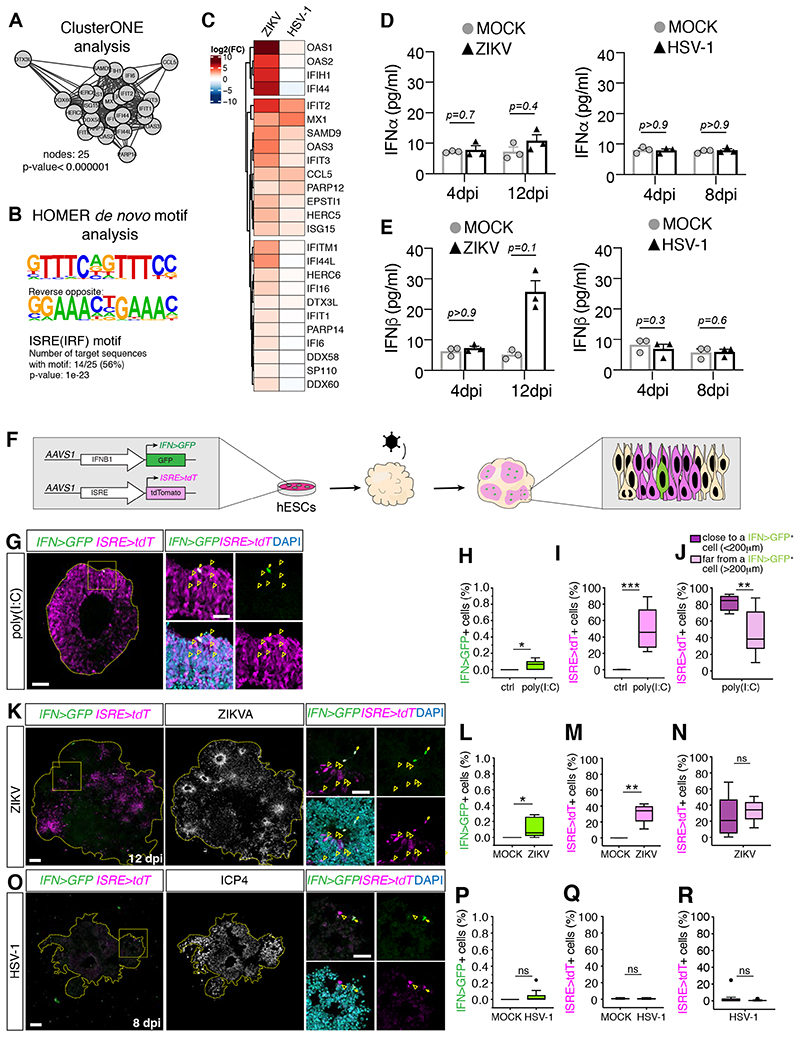
Figure 3. ZIKV and HSV-1 infections differentially engage the IFN-I system.
A-B) Cluster of densely connected genes among genes upregulated in ZIKV-infected organoids at 12 dpi (in A) and their HOMER de novo motif analysis (in B).
C) Expression analysis of genes from A in infected organoids compared to their MOCK controls. D-E) Quantification of IFNα and IFNβ levels measured by ELISA assay. Values represent mean ± SEM (Mann-Whitney tests).
F) Schematic diagram of IFNB1>GFP (IFN>GFP) and ISRE>tdTomato (ISRE>tdT) reporters.
G-R) Immunostaining (scale bars 100 μm) of organoids generated from reporter cells and analyzed one day after stimulation with poly(I:C) in G, or after ZIKV or HSV-1 exposure (in K and O). Dashed lines indicate organoid contour based on DAPI (not shown). Insets (scale bars 50 μm) represent a magnified view of the area close to one single IFN>GFP+ cell (arrows). Arrowheads indicate ISRE>tdT+ cells. Graphs are Tukey plots (n≥3; p=0.0211 in H, p=0.0007 in I, p=0.0068 in J, p=0.0273 in L, p=0.0091 in M, p=0.3175 in N, p=0.0857 in P, p=0.6820 in Q, p=0.5994 in R, Mann-Whitney tests). Ctrl, control transfection. ns, non-significant.
dpi, days post-infection; See also Figure S3.