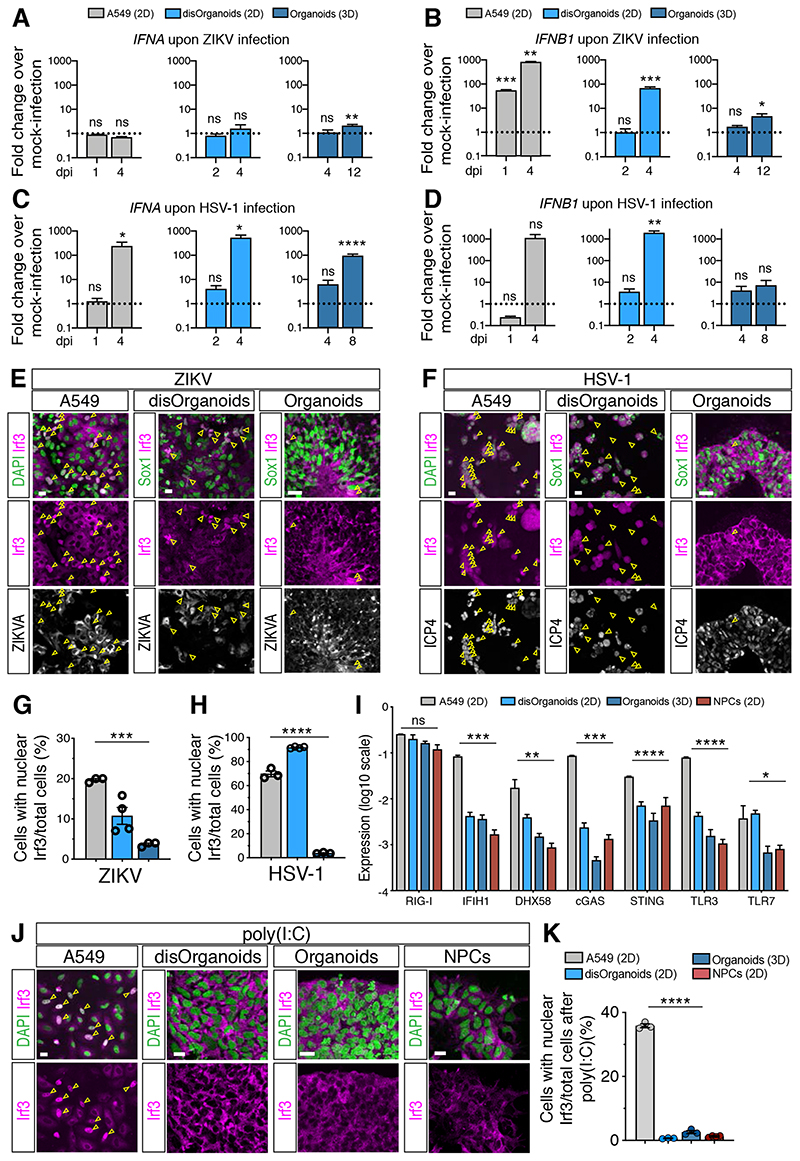
Figure 4. The IFN-I response in brain organoids is more attenuated than in 2D cultures.
A-D) Quantification of IFNA and IFNB1 expression by RT-qPCR in cultures exposed to ZIKV (in A-B) or HSV-1 (in C-D). Dotted lines indicate the value of 1. Values are mean ± SEM (n=3 for A549, n=4 for disOrg, n=7 for ZIKV organoids, n=3 for HSV-1 organoids; ZIKV IFNA: p=0.8985 A549 1 dpi; p=0.0823 A549 4 dpi; p=0.6069 disOrg 2 dpi; p=0.9784 disOrg 4 dpi; p=0.7633 Org 4 dpi; p=0.0057 Org 12 dpi; ZIKV IFNB1: p=0.0005 A549 1 dpi; p=0.0015 A549 4 dpi; p=0.7036 disOrg 2 dpi; p=0.0008 disOrg 4 dpi; p=0.2135 Org 4 dpi; p=0.0333 Org 12 dpi; HSV-1 IFNA: p=0.5876 A549 1 dpi; p=0.0105 A549 4 dpi; p=0.1457 disOrg 2 dpi; p=0.0183 disOrg 4 dpi; p=0.2903 Org 4 dpi; p<0.0001 Org 8 dpi; HSV-1 IFNB1: p=0.0794 A549 1 dpi; p=0.1054 A549 4 dpi; p=0.3126 disOrg 2 dpi; p=0.0056 disOrg 4 dpi; p=0.4055 Org 4 dpi; p=0.2250 Org 8 dpi; Mann-Whitney test comparisons of infected samples vs age-matched MOCK samples).
E-H) Immunostaining (scale bars 100 μm) and quantification of Irf3 nuclear localization. A549 and disOrganoids were analyzed at 4 dpi, organoids at 12 dpi (in E) or 8 dpi (in F). ZIKVA, Zika virus Antigen; ICP4, infected cell polypeptide 4 protein of HSV-1. Color code as in A. Values are mean ± SEM (p=0.0007 in G; p<0.0001 in H; one-way ANOVA).
I) Expression of nucleic acid sensors measured by RT-qPCR. Values are mean ± SEM (n=3, p=0.3387 RIG-I; p=0.0063 DHX58; p=0.0432 TLR7; **** is p<0.0001; one-way ANOVA).
J-K) Immunostaining (scale bars 20 μm) and quantification of Irf3 nuclear accumulation after poly(I:C) treatment. Values are mean ± SEM (n=3; p<0.0001, one-way ANOVA).
ns, non-significant; dpi, days post-infection. See also Figure S4.