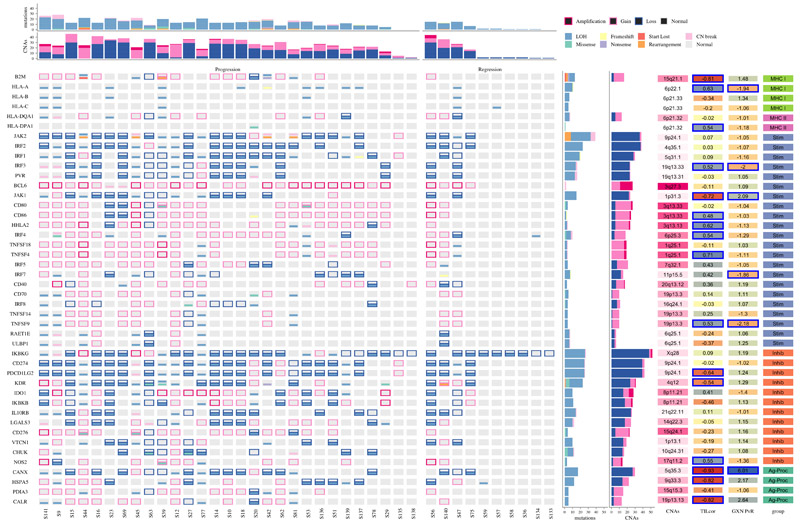
Figure 3. Genomic aberrations affecting immune genes in lung carcinoma in-situ lesions.
The mutational status is shown for 62 genes involved in the immune response, which are expressed by antigen presenting (tumor) cells. Genes are categorized as belonging to the Major Histocompatibility Complex (MHC) class I or II; stimulators (Stim) and inhibitors (Inhib) of the immune response, and genes involved in antigen processing (Ag-Proc). Mutations and copy number aberrations (CNAs) are shown for each of 29 progressive and 10 regressive samples. Loss of heterozygosity (LOH) events are shown as mutations to avoid confusion with copy number loss, relative to ploidy. The GXN PvR column displays the fold-change in expression of each gene between progressive and regressive samples, defined in a partially overlapping set of 18 samples. Significant genes, defined as False Discovery Rate < 0.05, are highlighted in blue. The TILcor column displays the Pearson’s correlation coefficient between the expression of each gene and the gene-expression based tumour infiltrating lymphocyte (TIL) score, derived by the Danaher method. Progressive samples had more mutations (p=0.028) and CNAs (p=0.0038) than regressive in this gene set. dN/dS analysis identified B2M, CHUK, KDR and CD80 as showing evidence of selection.