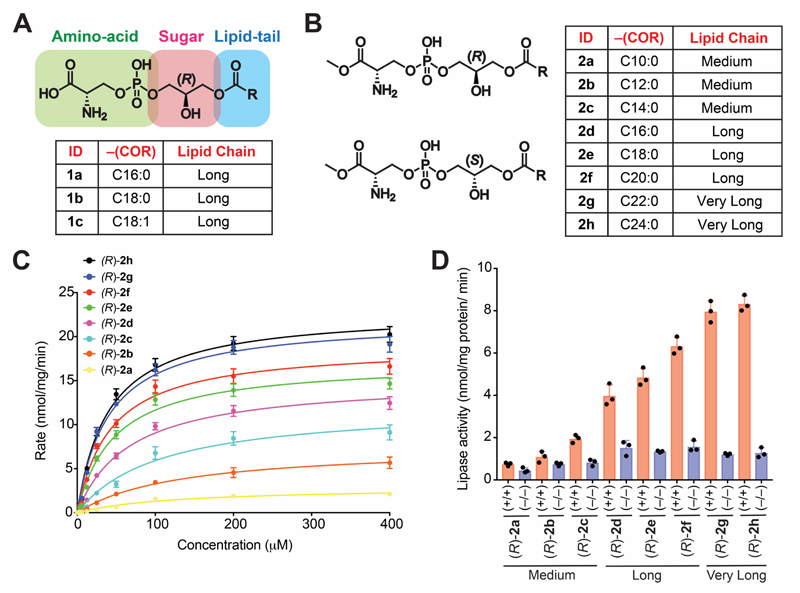
Figure 1. Structure of Me-lyso-PSs, and testing them as substrates against ABHD12.
The chemical structures of (A) commercially available canonical natural lyso-PSs and (B) our synthetic Me-lyso-PS lipid library with both (R)- and (S)-stereoisomers. (C) Enzyme kinetic assays for membrane lysates (10 㭜g) of HEK293T cells transfected with hABHD12 tested against the (R)-Me-lyso-PSs (0 – 400 㭜M, 30 mins, 37 °C, n = 3/data point). The line connecting the points, represents a fit to the Michaelis-Menten enzyme kinetics equation. See Table 1 for all enzyme kinetics parameters. (D) Lipase assays for (R)-Me-lyso-PSs (100 㭜M, 30 mins, 37 °C, n = 3/group) tested against brain membrane lysates (20 㭜g) from wild type (+/+) or ABHD12 knockout (–/–) mice. All data presented in (C, data points), and (D, bars) is represented as mean ± standard deviation.