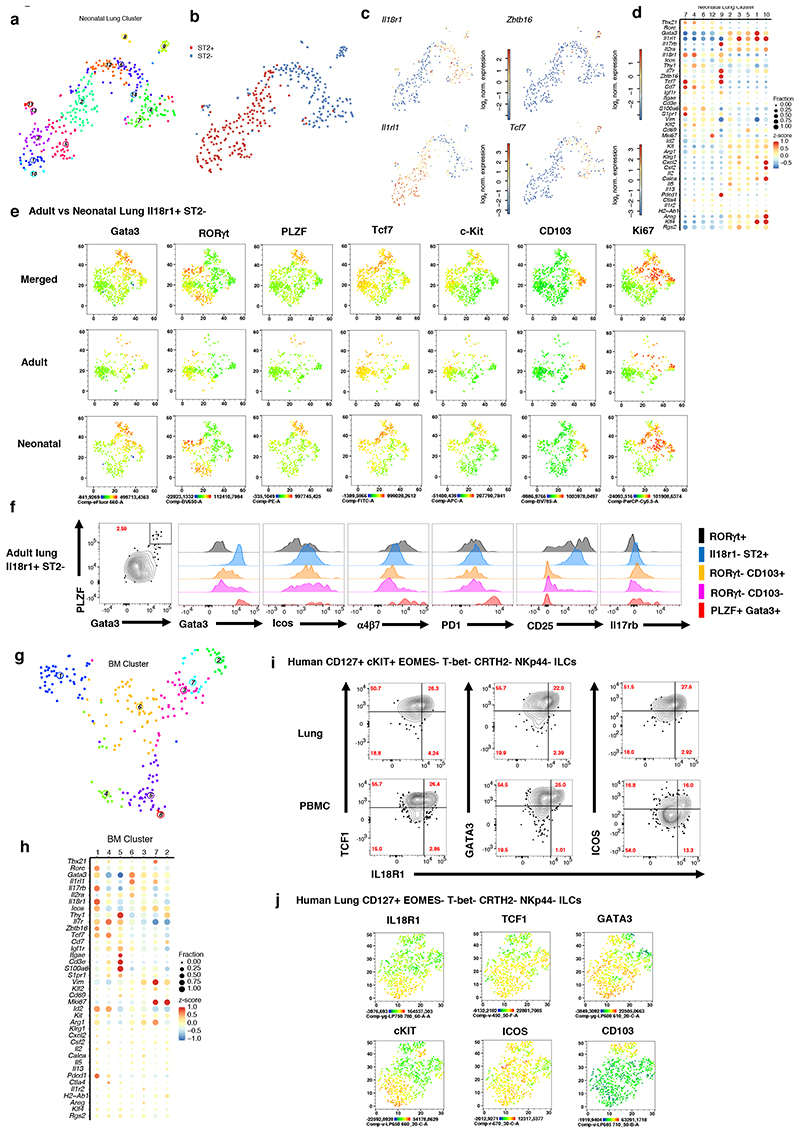
Figure 2. IL18rl+ST2- ILCs in neonatal and adult lung share similarities with IL18rl+ BM ILCPs.
(A-C), t-SNE maps of neonatal lung ILC single-cell transcriptomes highlighting RaceID3 clusters (A) or sorted ST2- and ST2+ ILCs (B), and log2 normalized expression of representative genes marking putative ILC progenitors (C). (D) Expression of candidate genes within neonatal lung clusters with at least 10 cells. Color represents z-score mean expression across clusters and dot size represents fraction of cells in the cluster expressing the gene. (E) FACS analysis of neonatal (P4) and adult (6-8w) Il18r1+ST2- lung ILCs, represented as t-SNE maps, indicating expression levels for Gata3, RORγt, PLZF, Tcf7, C-kit, CD103 and Ki67. (F) Histograms comparing ILC subsets (PLZF+Gata3+, RORγt+CD103-, RORγt+CD103+, Il18r1-ST2- ILC2s and RORγt+ ILC3) in adult lung for Gata3, Icos, a4p7, PD1, CD25 and Il17rb expression levels. Data are representative of 2-3 independent experiments (n=6-12 mice). (G) t-SNE map of Il18r1+Icos+ BM ILCs highlighting RaceID3 clusters. (H) Expression of candidate genes for Il18r1+Icos+ BM clusters with at least 10 cells. Color represents z-score mean expression across clusters and dot size represents fraction of cells in the cluster expressing the gene. (I-J) FACS analysis of human lung and PB ILCs pre-gated on CD45+Lin- CD127+EOMES-T-bet-CRTH2-NKp44-c-KIT+/- (I) or c-KIT ILCs (J). Figures depict representative staining (I) and t-SNE maps (J) indicating the expression levels of selected markers. Data are representative of 3 independent experiments (n=2-4 samples each).