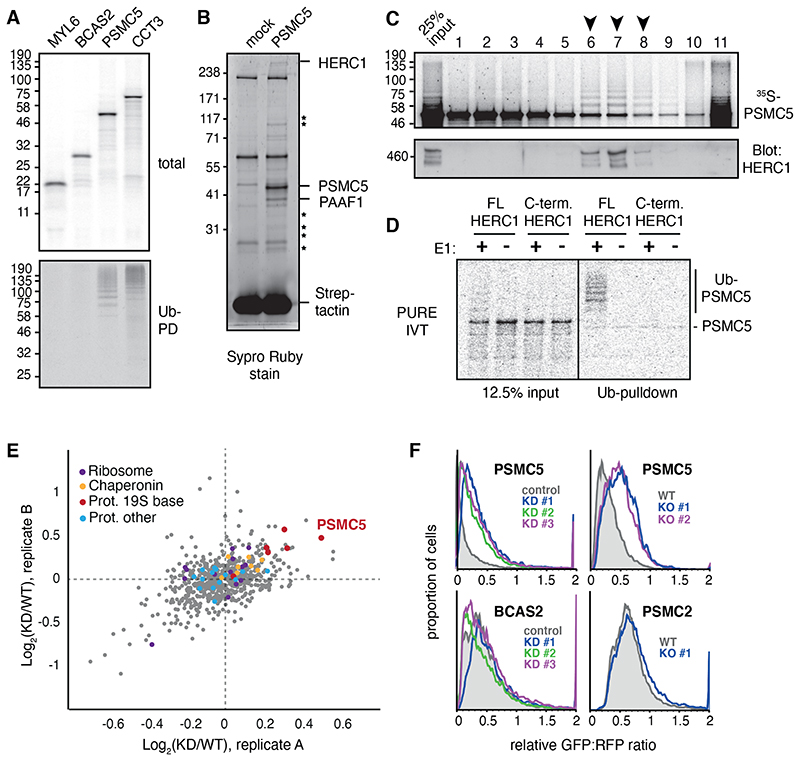
Fig 2. HERC1 mediates nascent PSMC5 degradation.
(A) Subunits of four different multimeric complexes were translated in RRL containing 35S-methionine and His-tagged ubiquitin (Ub). Ubiquitinated translation products were isolated by Ni-NTA pulldown under denaturing conditions. (B) PSMC5 containing a twin-Strep tag (TST) was translated in RRL, affinity purified under native conditions using immobilized Streptactin, and the products visualized by SYPRO Ruby stain. Mock indicates a parallel reaction lacking mRNA. The indicated PSMC5-specific interaction partners were identified by mass spectrometry, with stars denoting other 19S subunits of the proteasome. (C) 35S-methionine labelled PSMC5 translated in in RRL was separated on a 5-25% sucrose gradient. Fractions were analysed by autoradiography for PSMC5, and by immunoblot for HERC1. Arrows indicate fractions where ubiquitinated PSMC5 co-factionates with HERC1. On this gradient, ~300 kD protein typically migrates in fractions 3 to 5, and a ~500 kD protein in fractions 5 to 7. (D) 35S-labeled PSMC5 in complex with PAAF1 was produced by translation in the PURE system (see fig. S5A). The complex was mixed with E1, E2, His-Ub, ATP, and either full length or truncated recombinant HERC1 (see fig. S5B). Ubiquitinated products were isolated by Ni-NTA pulldown under denaturing conditions and visualized by autoradiography. (E) MCF7 cells were pretreated with nontargeting (WT) or HERC1-targeting (KD) siRNAs and metabolically labeled for 1 hour with AHA. The labeled proteins were selectively recovered via click chemistry, and the recovered proteins were analyzed by quantitative mass spectrometry. The top 20% most abundant proteins from two biological replicates are plotted by the KD/WT ratio, with subunits of the proteasome, ribosome, and chaperonin highlighted. (F) Flow cytometry analysis of GFP-tagged reporter proteins compared to an internal RFP control. HERC1 was knocked down (KD) by 3 separate siRNAs or knocked out (KO) using CRISPR.