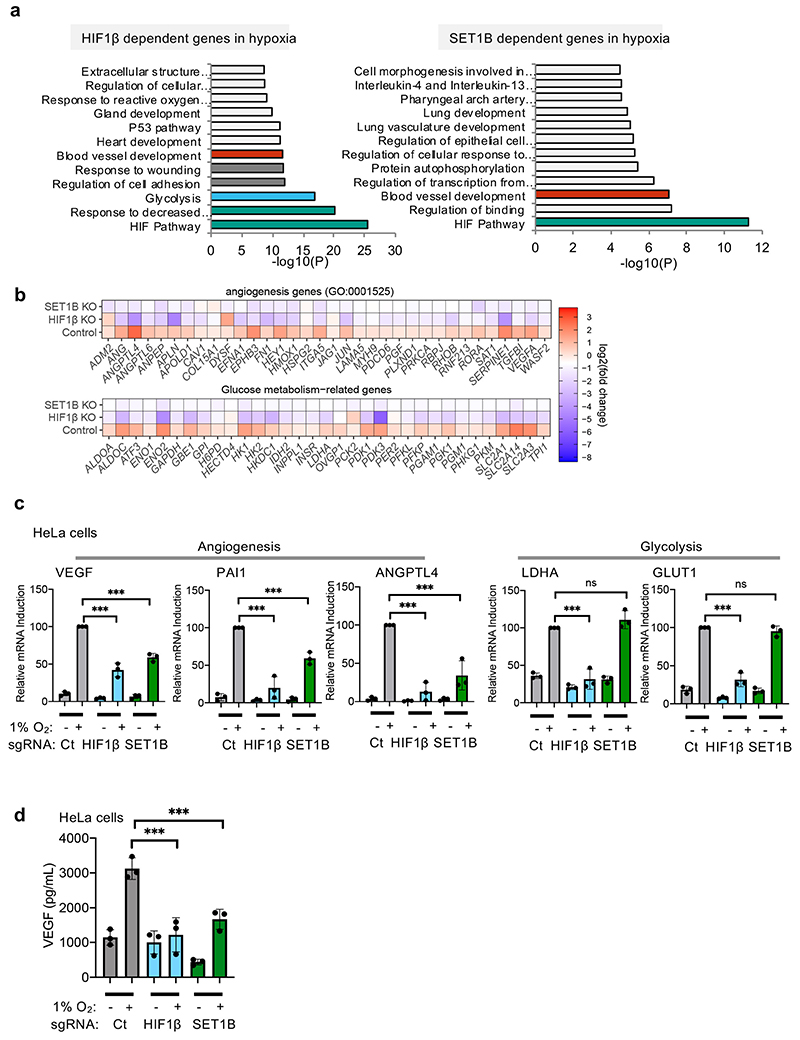
Extended Data Fig. 5. SET1B regulates the expression of a subset of HIF-target genes.
(a) Pathway analysis of enriched genes dependent on HIF1β or SET1B in hypoxia. HIF1β and SET1B dependent genes identified from the RNA Seq were run through Metascape analysis to identify the pathways which were enriched. (b) Heat map comparing the log2(fold change) between control, HIF1β and SET1B depleted cells focussing on genes involved in angiogenesis and glycolysis. (c) qPCR of selected angiogenesis (left) or glycolysis genes (right) in mixed KO populations of HIF1β or SET1B HeLa cells. VEGF, control vs. HIF1β or SET1B sgRNA P ≤ 0.0001; PAI, control vs. HIF1β or SET1B sgRNA P ≤ 0.0001; ANGPTL4, control vs. HIF1β or SET1B sgRNA P ≤ 0.0001; LDHA, control vs. HIF1β sgRNA P ≤ 0.0001; GLUT1, control vs HIF1β sgRNA P ≤ 0.0001; two-way ANOVA, n= 3 biological replicates. (d) VEGF ELISA in control, HIF1β or SET1B depleted HeLa cells. Cells were grown in 21% or 1% O2 before supernatants were collected. VEGF control vs. HIF1β or SET1B sgRNA P ≤ 0.0001; two-way ANOVA, n= 3 biological replicates. Ct=control. Graphs show mean ± SD, *** P < 0.001.