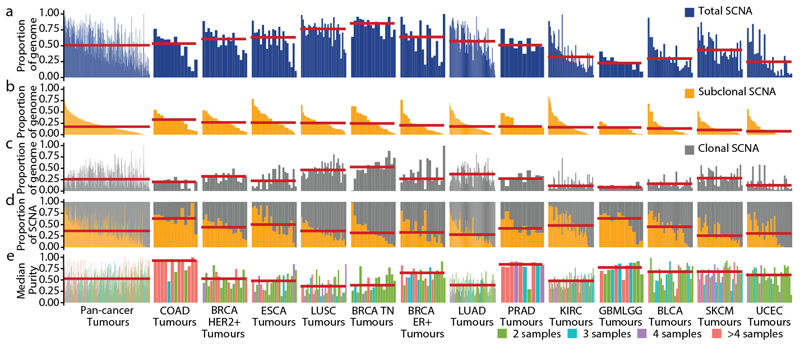
Fig. 1. Overview of somatic copy number heterogeneity across tumour types.
a, For each tumour, the proportion of the genome that is affected by SCNAs (both clonal and subclonal) is indicated. Tumour types with tumour samples from at least 10 patients were included: colorectal adenocarcinoma (COAD, n = 13), HER2+ breast cancer (HER2+ BRCA, n = 18), oesophageal adenocarcinoma (ESCA, n = 22), lung squamous cell carcinoma (LUSC,n = 31), triple-negative breast cancer (TN BRCA, n = 17), ER+ breast cancer (ER+ BRCA, n = 19), lung adenocarcinoma (LUAD, n = 84), prostate adenocarcinoma (PRAD, n = 10), clear cell renal cell carcinoma (KIRC, n = 54), glioma (n = 12), bladder urothelial carcinoma (BLCA, n = 26), melanoma (SKCM, n = 30) and endometrial carcinoma (UCEC, n = 27). Tumour types and tumours are ordered by the median proportion of the genome that is affected by subclonal SCNA—this order is maintained throughout the figure. Red lines indicate the median of the distribution. b, c, The proportion of the genome affected by subclonal (b) and clonal (c) SCNAs. d, The proportions of SCNAs that are subclonal and clonal are shown. The red line indicates the median proportion of SCNAs that are subclonal. e, The median purity and number of samples for each tumour.