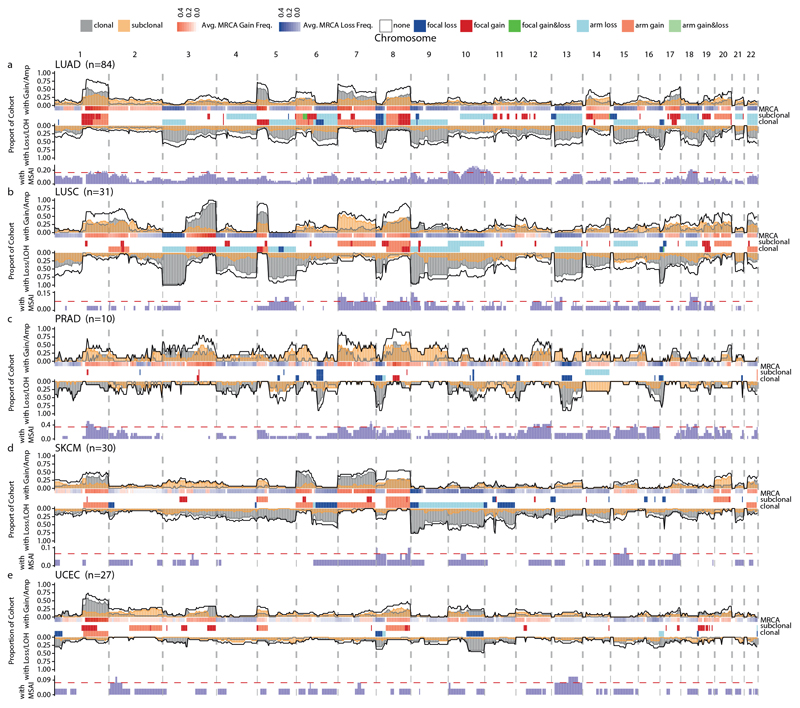
Extended Data Fig. 7. Subclonal SCNA landscape across tumour types.
a–e, The following tumour types were analysed: LUAD (a; n = 84), LUSC (b; n = 31), prostate adenocarcinoma (c; n = 10), SKCM (d; n = 30) and endometrial carcinoma (e; n = 27). Across-genome plots show clonal and subclonal SCNAs. Within each tumour type for each chromosome, the following data are shown (top to bottom): the proportion of patients with gains or amplifications. The black line indicates the total proportion of patients with gains/amplifications; the yellow and grey lines or shades indicate the proportion of patients with subclonal and clonal gains, respectively. The MRCA was derived by phylogenetic analysis (see Methods, ‘Ancestral reconstruction and phylogeny inference’). For each locus, the frequency of gains (red) and losses (blue) found in the MRCAs of the tumours are indicated. The GISTIC2.0 events. These tracks indicate significant SCNA focal events that were identified by GISTIC2.0 (see Methods, ‘GISTIC2.0 peak definition’ and ‘GISTIC2.0 consensus peak definition’) and recurrent arm-level events (see Methods, ‘Arm-level SCNA definition’). The proportion of patients with loss/LOH events. The black line indicates the total proportion of patients with loss/LOH events; the yellow and grey lines or shades indicate the proportion of patients with subclonal and clonal losses, respectively. The black, yellow and grey lines indicate significance thresholds for total loss/LOH, subclonal loss/LOH and clonal loss/LOH, respectively. Proportion of patients with mirrored subclonal allelic imbalance (MSAI) originating from distinct haplotypes identified by multi-sample phasing. The red line indicates the significance threshold determined by a permutation test at the 0.05 level (see Methods, ‘Permutation test for recurrence of SCNA across tumours’).