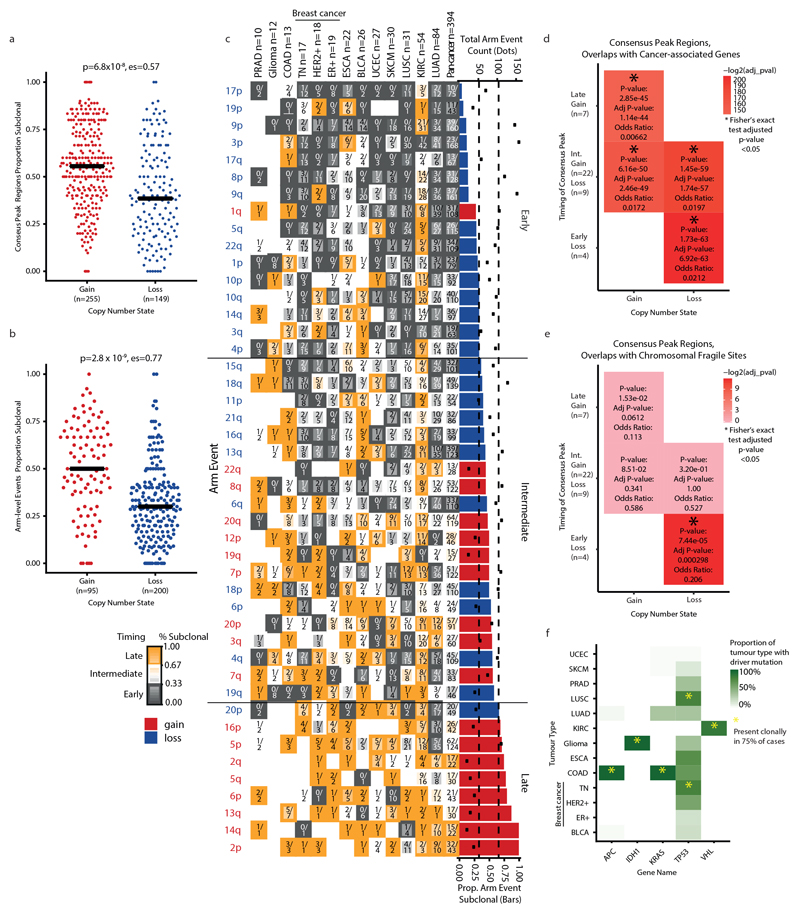
Extended Data Fig. 8. Recurrent SCNA across tumour types.
a, b, Difference in gains and losses in consensus-peak region gains (red, n = 255) and losses (blue, n = 149) (a) and chromosome arm gains (red, n = 95) and losses (blue, n = 200) across all tumour types (b). Black horizontal bars indicate the median of the distribution. Significance testing was performed using an unpaired Student’s t-test. c, Classification of chromosomal arm-level events according to timing. Left, heat map of the percentage of subclonal occurrence of all events in each tumour type. The numerator within each cell indicates, in that tumour type, the total number of subclonal occurrences of that event and the denominator indicates the total number of both clonal and subclonal occurrences of that event in that tumour type. Shading of each cell in the heat map indicates the percentage of subclonal occurrences of an event within a tumour type with orange indicating a higher subclonality and grey indicating a higher clonality. The border of each cell indicates the classification of that event in a tumour type as either early (grey border), intermediate (no border) or late (orange border). Right, bar plot of arm-level events ordered by median percentage of subclonal occurrences across tumour types (bottom axis). Bars representing gain events are coloured in red and loss events are coloured in blue. Horizontal black lines indicate separation of events into pan-cancer categories of early, intermediate and late, according to tertiles of the median proportion of SCNAs that is subclonal. Dots centred on the same axis positions indicate the total event count of each loss or gain event across tumour types (top axis). d, Enrichment of early, intermediate and late consensus peak events with known cancer-associated genes. Heat map indicating the resulting P values from two-sided Fisher’s exact tests comparing the overlap of genes in early, intermediate and late consensus peaks with previously reported oncogenes and tumour-suppressor genes. Gain peaks were investigated in relation to oncogenes, while loss peaks were investigated in relation to tumour-suppressor genes. Significant overlaps (Benjamini–Hochberg-adjusted P < 0.05) are indicated with an asterisk (see Methods, ‘Cancer-associated gene and fragile site enrichment’). e, Enrichment of early, intermediate and late consensus peak events with chromosome fragile sites. Heat map indicating the resulting P values from Fisher’s exact tests comparing the overlap of cytobands found in early, intermediate and late consensus peaks with cytobands from previously reported chromosome fragile sites. Significant overlaps (Benjamini–Hochberg-adjusted P < 0.05) are indicated with an asterisk (see Methods, ‘Cancer-associated gene and fragile site enrichment’). f, Prevalence of SNVs and indels in cancer-associated genes. Heat map displaying the proportion of samples from each tumour type with an SNV or indel in the corresponding cancer-associated gene. Yellow asterisks indicate where the SNVs and indels are present clonally in ≥75% of tumours in the corresponding tumour type.