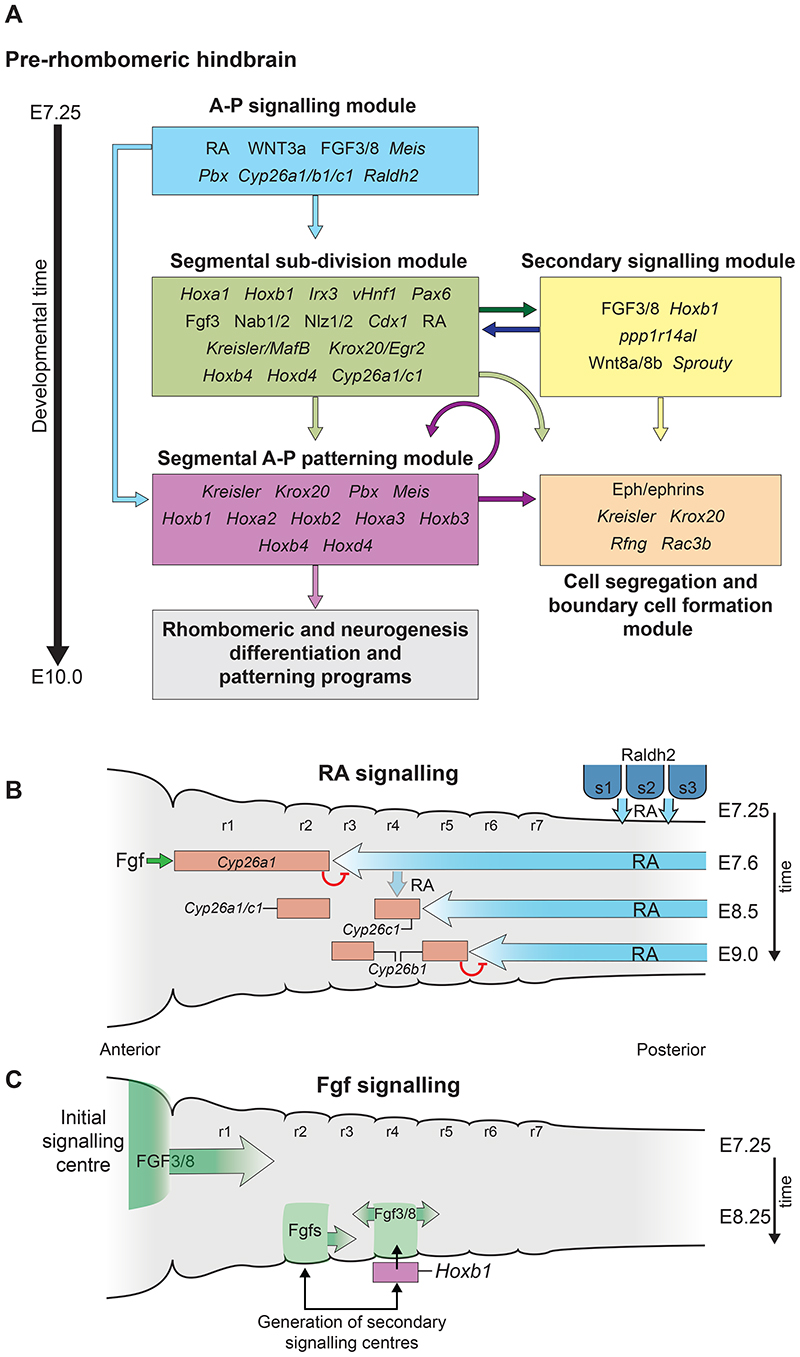
Fig. 2. Hierarchical structure of the gene regulatory network (GRN) governing hindbrain segmentation.
A framework of the GRN model depicted as a progressive series of modules/steps associated with underlying cell and developmental processes in the mouse hindbrain between E7.25 and E10.0. Each module (coloured boxes) has its own layer of regulatory circuits and components, as indicated by the key genes and signals inside each box. The gene names correspond to mouse and may vary in other vertebrates. The arrows indicate the flow of regulatory information between modules. Many genes and signals have fundamental roles in multiple steps of the segmentation and patterning process. Regulatory interactions in the A-P signalling module set up temporally and spatially dynamic domains of signalling. (B) Diagram of shifting RA gradients involved in regulating early events in mouse hindbrain segmentation in stages between E7.5 and E9.0. RA is generated by Raldh2 in somitic mesoderm and diffuses anteriorly in the hindbrain. Fgf signals from the midbrain isthmic organiser induce Cyp26a1 which degrades RA in r1 and r2. This establishes an initial RA signalling domain with an anterior limit at the future r2/3 border at E7.5. Cyp26c1 is expressed in r4 at E7.9, and slightly later (E8.5) Cyp26b1 is induced in r3 and r5. This progressive activation of Cyp26 genes (red rectangles), and degradation of RA, creates shifting gradients of RA (blue arrows) that eventually establish an anterior limit at the r5/6 boundary by E9.0. (C) Diagram of dynamics of initial and secondary Fgf signalling centres. At E7.25 an initial Ffg signalling centre forms at the isthmic organizer in the mid/hindbrain border region and patterns the anterior hindbrain. At E8.5 initiation of Hoxb1 in r4 leads to activation of Fgf3/8 and the formation of a secondary signalling centre in r4 important for patterning the posterior hindbrain. In chick embryos there is evidence for a secondary Fgf signalling centre in r2. Figure adapted from Fig. 2 and 3 of (Parker and Krumlauf, 2020). r, rhombomere; s, somite; RA, retinoic acid.