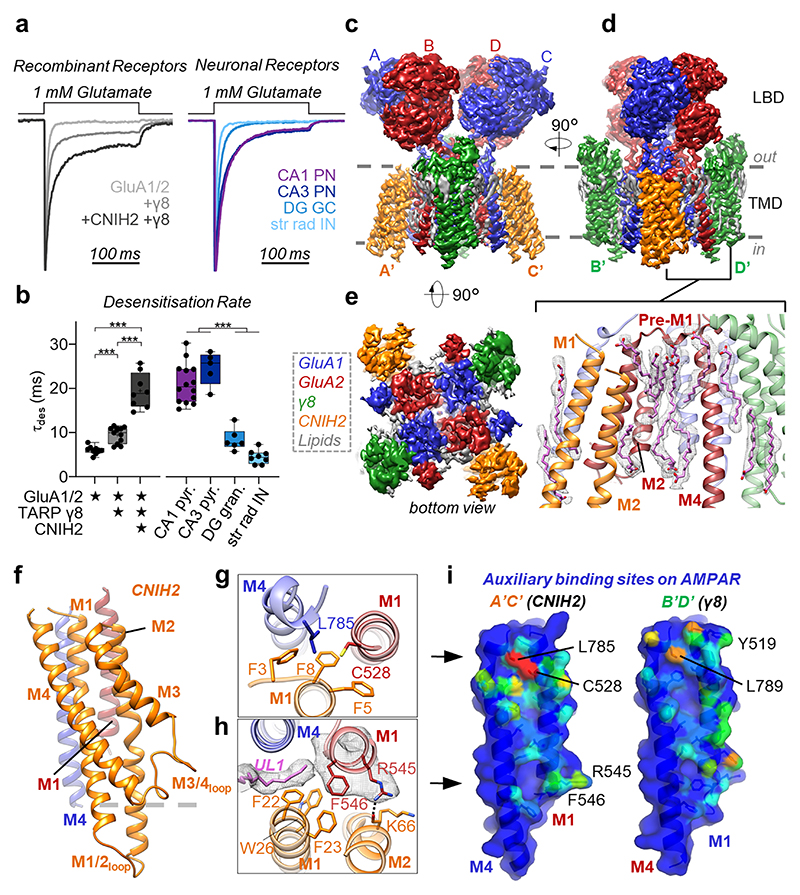
Fig. 1. Physiology and architecture of the GluA1/2_γ8 /CNIH2 complex.
a,b, Auxiliary subunits slow desensitization rates of recombinant receptors (a, left). Native AMPARs show diverse properties (a, right), with A1/2_γ8 +CNIH2 recapitulating CA1 & CA3-like kinetics (b, weighted τdes (ms), mean ± SEM). Recombinant receptors: GluA1/2: 6.01 ± 0.31, n=9; +γ8: 9.29 ± 0.53, n=12; +CNIH2+γ8: 19.71 ± 1.27, n=9. Neuronal receptors: CA1 pyramidal: 21.03 ± 1.19, n=14; CA3 pyramidal 24.51 ± 1.69, n=5; DG granule cell: 8.38 ± 1.01, n=6; CA1 stratum radiatum interneurons: 4.48 ± 0.55, n=8; Welch’s ANOVA tests with Dunnett’s multiple comparisons test - Recombinant: W(2,15.11) = 60.68, p<0.0001; Neurons: W(3,11.60) = 76.28, p<0.0001; see Supplementary Table 1 for details). Boxes represent 25 % to 75 % percentile, whiskers minimum/maximum values and central line median. c-e, Cryo-EM maps, depicting the LBD and TMD domain layers. Core subunits positioned to AC/BD, and auxiliary subunits at A’C’/B’D’ sites are shown. (c) Front view, depicting γ8 at the B’D’ sites. (d) Side view, visualising CNIH2 at A’C’ sites. Inset: lipids concentrating at the TMD, beneath the GluA2 pre-M1 helix. (e) Bottom view, highlighting CNIH2 binding to GluA2 transmembrane helices M1-3 (red), and γ8 to the GluA1 M1-3 helices (blue). f, Model of CNIH2, including the M1/2 and M3/4 cytosolic loops, docking to its binding site (M1GluA2 [red] M4GluA1 [blue]). g, CNIH2 Phe3, -5, -8 slotting into its binding site close to Cys528 (GluA2) and Leu785 (GluA1). h, CNIH2 contacts at the bottom of the binding site, mediated by Phe23 and Lys66; lipid (UL1) penetrates the A’C’ site and interacts with GluA2 Phe546. i, A’C’ (left) and B’D’ (right) site surface representation. M1A2 and M4A1 residues contacted by CNIH2 are coloured depending on the number of atoms contributing to the interaction (red: high, blue: low). Contacts were counted using ‘findNeighbors’ in ProDy38’, with a 4.5 Å cutoff between heavy atoms.