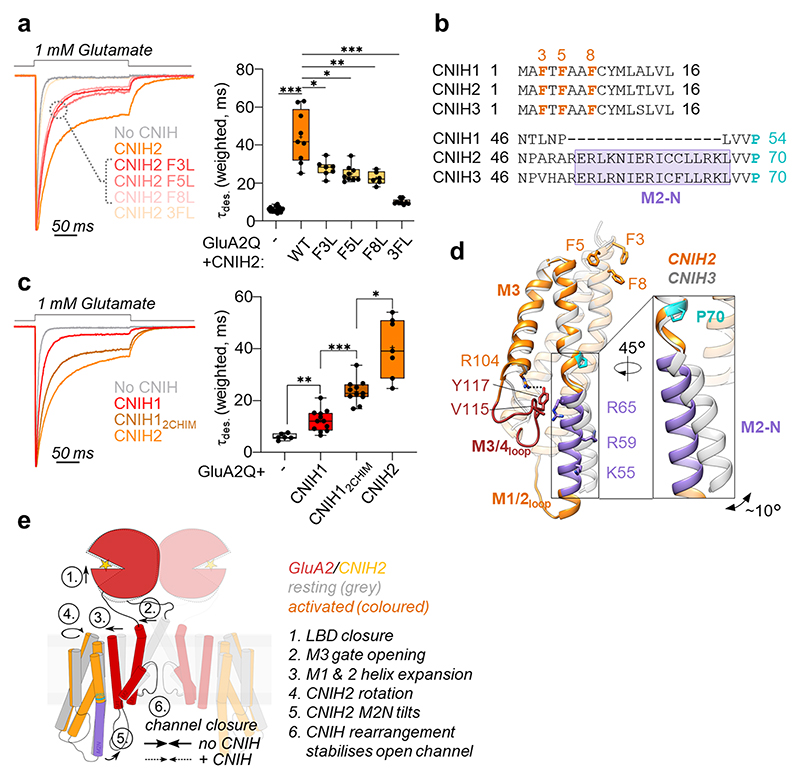
Fig. 3. Mechanism of AMPAR modulation by CNIH2.
a, Mutations of three phenylalanines in the CNIH2 N-terminus speed GluA2 desensitization: weighted τdes (ms), mean ± SEM – GluA2 alone: 6.16 ± 0.35, n=15; CNIH2: 44.32 ± 4.58, n=9; F3L: 27.73 ± 1.58, n=7; F5L: 24.85 ± 1.43, n=9; F8L: 22.57 ± 1.40, n=6; 3FL: 10.07 ± 0.46, n=9; Welch’s ANOVA with Dunnett’s multiple comparisons test: W(5,17.83) = 85, p<0.0001; further details in Supplementary Table 2. Boxes represent 25 % to 75 % percentile, whiskers minimum/maximum values and central line median. b, Sequence alignment of mouse CNIH1-3, highlighting the conserved N-terminal phenylalanines and the M2-N region, where CNIH1 lacks 16 residues; Pro70 is shown in cyan. c, The CNIH2 M2-N helix contributes to modulation of GluA2 kinetics, demonstrated by a gain-of-function when transplanted onto CNIH1: weighted τdes (ms), mean ± SEM – GluA2 alone: 6.03 ± 0.47, n=6; CNIH1: 12.22 ± 1.20, n=12; CNIH12chimera: 23.48 ± 1.39, n=11; CNIH2: 40.45 ± 4.32, n=7; Welch’s ANOVA with Dunnett’s multiple comparisons test: W(3,15.30) = 64.09, p<0.0001; further details in Supplementary Table 2. Box plot parameters as in b. d, Superposition of CNIH2 (orange) and CNIH3 (grey; PDB 6PEQ), using the top of their M1 and M2 helices. CNIH2 M2-N (purple) kinks at Pro70 (cyan) and diverges from CNIH3 by ~ 10° (inset); the N-terminal phenylalanines are also indicated. An interaction with the M3/4 loop through Val115 and Arg65 is shown, Arg55 and Arg59 project toward the pore axis. e, Molecular mechanism underlying AMPAR modulation by CNIH2.