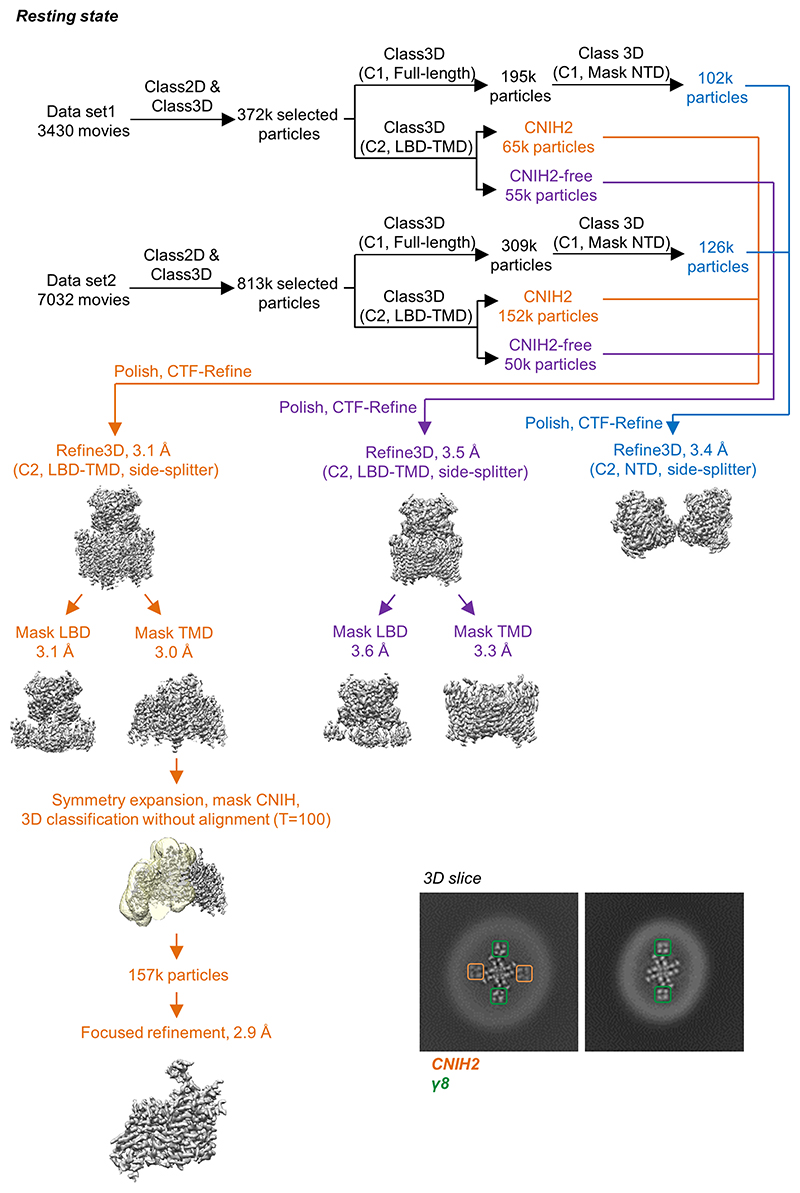
Extended Data Figure 2. Cryo-EM data processing workflow of the resting state A1/2_γ8/C2 complex.
Two datasets were first processed individually to remove particles lacking AMPAR features. Next, classifications focused on the LBD-TMD region were performed to separate CNIH-containing and CNIH-free particles, meanwhile classifications for full-length receptors were conducted to elucidate particles with a stable NTD signal. Subsequently, particles from the two datasets were combined together for refinement. Focused refinements were performed separately on the LBD-TMD gating core and the NTD region. To further improve the resolution, LBD and TMD are refined separately A structure of A1/2_γ8 (lacking CNIH2) was also resolved from the same dataset (containing only γ8 observed in 3D slice). CNIH density was further enhanced by applying first symmetry expansion on aligned particles from the TMD reconstruction, following by focused classification and refinement on only CNIH2 and the surrounding receptor transmembrane helices. Inset: Top view slices of the A1/2_γ8/C2 (left) and A1/2_γ8 (right). 3D maps at the TMD region show signal for transmembrane helices of γ8 (green) and CNIH2 (orange).