Abstract
Early prediction of risk of cardiovascular disease (CVD) including stroke, is a cornerstone of disease prevention. Clinical risk scores have been widely used for predicting CVD risk from known risk factors. Most CVDs have a substantial genetic component, which also has been confirmed for stroke in recent gene discovery efforts. However, the role of genetics in prediction of risk of CVD including stroke, has been limited to testing for highly penetrant monogenic disorders. In contrast, the importance of polygenic variation, the aggregated effect of many common genetic variants across the genome with individually small effects, has become more apparent in the last 5–10 years and powerful polygenic risk scores for CVD have been developed. Here we review the current state of the field of polygenic risk scores for CVD including stroke, and their potential to improve CVD risk prediction. We present findings and lessons from diseases such as coronary artery disease as these will likely be useful to inform future research in stroke polygenic risk prediction.
Non-standard Abbreviations and Acronyms
- PRS
polygenic risk score
- metaGRS
meta genomic risk score
- GPS
genome-wide polygenic score
- PCE
pooled cohort equations
- C-index
concordance index
CVD risk prediction
Risk prediction is one of the key tools in primary prevention of CVD and stroke1, with the first risk calculators being based on the Framingham study2, 3, followed by the Pooled Cohort Equations4, QRISK35, SCORE6, and others. Risk calculators rely on risk factors such as age, sex, total and/or LDL cholesterol, systolic blood pressure, and diabetes, to predict the future risk of CVD (typically within 10 years). Individuals with high risk of CVD and stroke can be prioritised for lifestyle interventions and medications such as statins, antiplatelet, and antihypertensive treatments.
Despite advances in risk prediction, for many people the first sign of CVD risk is an event such as a stroke or myocardial infarction. Some of the reasons for this are: (i) lack of routine screening in the general population for the risk factors included in the calculators; (ii) a large number of individuals fall within intermediate risk categories, a group in which many events will occur; (iii) imperfect effectiveness and uptake of interventions or lifestyle modifications; (iv) most traditional risk factors are less useful early in life to predict risk later in life; (v) standard risk calculators do not capture all the risk of CVD, and there remains substantial unexplained variability in risk, which may be partially explained by other factors not yet routinely incorporated into these scores, such as education levels7.
With the aim of identifying further biomarkers and risk factors to add information to clinical risk factors, genetics seems like a natural avenue, as there is the potential to discover new markers beyond the traditional risk factors, as well as take advantage of the fact that germline variation is essentially fixed for life and thus less susceptible to confounding and reverse causality than other biomarkers and risk factors.
Monogenic variation (also called Mendelian) and its effect on CVD risk has been well studied across a range of diseases: mutations for familial hypercholesterolemia (LDLR / APOB / PCSK9) increase the risk of myocardial infarction 4–10x fold8, and similarly Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy (CADASIL; NOTCH3) increases risk of ischaemic stroke markedly9.
While monogenic variants can have high penetrance for disease and are thus of high importance for those individuals who carry them and their families, only a small proportion of the population carry these risk variants (~1 in 250 for familial hypercholesterolemia, 2–4 in 100,000 for CADASIL10), and the vast majority of CVD events in the general population are not caused by them.
From GWAS to Polygenic Risk Scores
In contrast with rare monogenic variants, the role of common genetic variants in complex diseases (typically single nucleotide polymorphisms, SNPs, with minor allele frequency of >5%) has become clearer with the genetic and genomic revolution of the past 15 years or so. Genome-wide associations studies (GWAS) have now identified as many as 32 genetic loci associated with stroke11, >160 loci associated with coronary artery disease12–14, and other related traits such as intracranial aneurysm15 (17 loci) and blood pressure16–19 (>1000 loci). The heritability of CAD and stroke has been estimated at ~40–50%20. However, despite the large number of risk loci identified so far, these typically still explain only a minority of the heritability of CVD and stroke (~20% for CAD and ~2% for ischaemic stroke), indicating that there are likely more loci yet to be discovered by further increasing the GWAS sample size21 and examining SNPs with lower allele frequencies. In addition, some variants may be ancestry-specific, discoverable only by analysis of populations from more diverse ancestries22.
The main aim of GWAS is biological discovery, but the results can also be harnessed for risk prediction via polygenic risk scores (PRS), also called genetic or genomic risk scores (GRS) or genome-wide polygenic scores (GPS). While the individual alleles identified in GWAS typically modify the disease risks only marginally, the cumulative impact of risk alleles across the entire genome is considerable for many diseases. PRS are the weighted sum of the individual effects of many risk alleles, with the weight based on the effect size estimated from GWAS and optionally re-estimated to account for other properties such as linkage disequilibrium.
Early risk scores had only a small number of SNPs23–26, as they were based on genome-wide significant SNPs (typically SNPs associated at P<5x10-8, the genome-wide statistical significance threshold). Subsequent relaxing of this stringent requirement led to stronger yet still robust scores27–30. More recently, it has been demonstrated that expanding the scores to include far more SNPs than the genome-wide significant SNPs can substantially improve predictive power, in accordance with the high polygenicity of CVD. PRS with as many as several thousand or even millions of SNPs have been developed for CAD 31–33, atrial fibrillation32–34 and ischaemic stroke35, using large biobank studies including the UK Biobank36, 37 and FinnGen.
For ischaemic stroke (IS), expanding the PRS from a 90-SNP score27 (http://www.pgscatalog.org/score/PGS000038) to a 3.2 million SNP score (metaGRSstroke; http://www.pgscatalog.org/score/PGS000039)35 increased the association with ischaemic stroke from a hazard ratio (HR) of 1.13 (95% CI 1.10–1.17) per standard deviation of the score to HR=1.26 (95% CI 1.22–1.31) per standard deviation, and correspondingly increased the C-index by 0.029 (C-indices of 0.556 and 0.585 for the 90-SNP score and metaGRS, respectively; based on sex-stratified age-as-time-scale Cox models, adjusting for genotyping chip and 10 genetic principal components). Another PRS, consisting of 350,000 SNPs, was recently developed based on Japanese case/control datasets including BioBank Japan, and validated in the Hisayama Study in Japan, resulting in a sex and age-adjusted HR=1.64 (95% CI 1.04–2.55) when comparing the bottom 40% with the top 60% of the score38.
For coronary artery disease, an analysis in the UK Biobank31 showed that an early 50-SNP score (http://www.pgscatalog.org/score/PGS000011)25 achieved a hazard ratio of 1.26 (95% CI 1.25–1.28) per standard deviation (SD) of the score, compared with HR=1.52 (95% CI 1.50–1.55) for a later 46,000-SNP score28 (http://www.pgscatalog.org/score/PGS000012), and increasing to HR=1.71 (95% CI 1.68–1.73) for a 1.7 million SNP score (metaGRSCAD; http://www.pgscatalog.org/score/PGS000018). The C-index (discrimination) increased from C=0.59 (95% CI 0–58–0.60) for the 50-SNP score to C=0.66 (95% CI 0.65–0.66) for the metaGRSCAD. Similar results in the UK Biobank were achieved using an alternative approach based on a PRS consisting of 6.6 million SNPs (GPSCAD; http://www.pgscatalog.org/score/PGS000013)32.
Despite early concerns that the use of SNPs that had not reached genome-wide significance in the construction of a PRS could reduce the reliability of such scores, the performance of these scores has been strongly validated in independent cohorts outside of UK Biobank: in a meta-analysis of French Canadian cohorts (MHI Biobank / CARTaGENE)39 the metaGRSCAD had an odds ratio (OR) of 1.69 (95% CI 1.58–1.81) per SD of the score for prevalent CAD and OR=1.17 (95% CI, 1.08–1.26) per SD of the score for recurrent CAD; the GPSCAD had an OR=1.61 (95% CI 1.51–1.71) per SD of the score for prevalent CAD and OR=1.13 (95% CI 1.06–1.22) per SD of the score for recurrent CAD. Similarly, in an analysis of incident- only CAD in the eMERGE cohort in the USA40, consistent effect patterns in individuals of European ancestries were observed: HR=1.20 (95% CI 1.15–1.25) per standard deviation for the 50-SNP score, HR=1.50 (95% CI 1.43–1.56) for the GPSCAD, and HR=1.53 (95% CI 1.46–1.60) for the metaGRSCAD, with both larger scores showing increases of C-index by +0.02 over the smaller score.
It is now clear that due to the high polygenicity of CVD, scores that incorporate a much larger number of SNPs than the genome-wide significant SNPs, can achieve substantially better predictive power with consistent performance across cohorts of the same ancestry. However, this approach also depends on the availability of more sophisticated statistical modelling tools that can better account for the complex patterns of SNPs correlations (linkage disequilibrium, LD) and effect sizes, such as LDpred41. In addition, extra predictive power can often be gained by leveraging GWAS of multiple correlated risk factors and diseases related to CVD and stroke (e.g., blood pressure, HDL cholesterol, BMI), into a single PRS, a so-called ‘metaGRS’31, 35.
PRS versus traditional risk factors
Genetic variation cannot directly affect disease risk but must operate through various biological pathways, some of which are represented by traditional risk factors such as blood pressure and cholesterol. The question of the relationship between PRS and traditional CVD risk factors has been examined across several studies with remarkably consistent results. More specifically, the questions of interest have been: (i) how does the predictive power of PRS compare with individual known risk factors? (ii) are PRS substantially correlated with known risk factors or independent of them? (iii) do PRS add any predictive information on top of established clinical risk scores? (iv) how do the unique characteristics of PRS affect risk prediction compared with that based on traditional risk factors?
First, recent CVD PRS have been shown to perform as well as or better than individual traditional risk factors. The metaGRSstroke has been shown in the UK Biobank to be more predictive of ischaemic stroke risk than family history, systolic blood pressure, body mass index (BMI), current smoking, diagnosed diabetes, and only slightly less than diagnosed hypertension35. Similarly for CAD, the metaGRSCAD has been shown in the UK Biobank to be more predictive of CAD risk than individual traditional risk factors (current smoking, diagnosed diabetes, family history of heart disease, hypertension, and diagnosed high cholesterol)31, with an increase in C-index of +0.029 for the PRS over the 2nd strongest risk factor, diagnosed high cholesterol (C-indices of 0.623 and 0.594 for the metaGRSCAD and diagnosed high cholesterol, respectively). Highly concordant results were demonstrated for the GPSCAD, which was shown to be stronger than each of the eleven CVD risk factors examined, including systolic and diastolic blood pressure, ApoB, ApoA1, total/LDL/HDL cholesterol, BMI, current smoking, diabetes, and family history of CAD, in both the Malmö Diet and Cancer Cardiovascular Cohort as well as the UK Biobank (increases in C-index of +0.013 and +0.014, respectively, over the next strongest factor, while adjusting for sex and age)42.
Second, across several studies, PRS for CVD have been shown to be essentially independent of the clinical risk scores such as the Framingham Risk Score and the Pooled Cohort Equations (PCE)28, 31, 33, 42. This is likely because age and sex are the largest contributors to these clinical scores43, whereas the PRS do not vary by sex or age. In addition, GWAS have uncovered a large number of genetic associations which strongly implicate the role of novel biological pathways not currently captured by traditional risk factors. In CAD, GWAS has highlighted the role of inflammation and cellular proliferation in disease risk44. Similarly in stroke, large meta-analyses have confirmed known associations with blood pressure, atrial fibrillation, and blood lipids as well as implicated pathways that are less of a focus of current research: coagulation, cardiomyocyte differentiation, muscle-cell fate commitment, nitric oxide metabolism, and possibly heart rate11. Since PRS that are based on genome-wide summary statistics are relatively unbiased with respect to biological pathways, they tend to capture a broad range of genetic effects which operate through traditional risk factors, novel risk factors, and those likely unknown as well. This, together with the fact that PRS act largely multiplicatively (log-additively) on disease risk, means that the PRS effect on risk is largely not explained by traditional risk factors and can be added into risk models as an essentially independent risk factor.
Third, beyond adding information above individual risk factors, recent CVD PRS also consistently increase predictive power above established clinical risk scores: adding the 49,000-SNP CAD PRS to either the Framingham Risk Score or the PCE led to increases in C-index for CAD of +0.015–0.016 (meta-analysis across FINRISK and Framingham Heart Study)28. Adding the GPSCAD to the PCE resulted in increases in C-index for CAD of +0.02 in UK Biobank and +0.026 in the Malmö Diet and Cancer Cardiovascular Cohort42. Another recent CAD PRS45 evaluated in the FINRISK study in Finland showed that a score based on age, sex, and a PRS, outperformed the clinical risk scores for predicting incident CAD (C-index of 0.832 and 0.823, respectively) and incident atrial fibrillation (C-index of 0.751 and 0.725, respectively).
Beyond the increases in raw predictive power, a public health analysis46 used the UK Biobank and extrapolated it to the general UK population via the Clinical Practice Research Datalink (CPRD) dataset. In the analysis, addition of a CVD PRS to conventional risk prediction models improved prediction of composite CVD (CAD or stroke). Targeting PRS to those individuals who had been identified as intermediate risk by clinical risk scores (5–10% 10-year risk) was estimated to help prevent one additional CVD event for every ~340 individuals screened. This compared favourably with an alternative approach of adding C-reactive protein (CRP) to the clinical models, which was estimated to lead to prevention of only one event in ~490 individuals screened. Extrapolating these estimates to the entire UK population, adding a PRS to risk screening would be estimated to prevent an additional 20,000 CVD events over 10 years in 40–75 year-old individuals.
Similarly, a health-economic analysis of the use of an earlier PRS28 to guide statin treatment for prevention of CAD found a net benefit in the context of the Finnish health system, with an estimated saving of 2.5 Euro per patient aged 45 or older, over a 10-year follow-up period47; it is likely that the net benefit for more recent CAD PRS, which are more powerful, would be even greater.
Finally, while PRS can be considered a powerful risk factor, it does offer unique advantages relative to traditional risk factors. Risk factors such as blood pressure and cholesterol levels can vary over time, and any one measurement only provides a snapshot at one point in time rather than the cumulative lifetime effect48. Similarly, family history often indicates both genetic as well as environmental/lifestyle risk, but is also highly limited in that it depends on actual events having occurred and being appropriately recorded and known by the person. In contrast, PRS are based on germline variation which is essentially stable across life and can be ascertained once by genotyping. This does not preclude updating the SNP weights when new scores become available in the future.
There is now strong evidence for creating new CVD risk scores, which combine traditional risk factors with PRS, leading to stronger scores than would be possible today. At the same time, the important role of monogenic variants should not be ignored. Since the polygenic and monogenic components are inherited essentially independently of each other, there will be individuals who carry monogenic variants as well as high PRS, putting them at substantially high risk of CVD versus carriers with low or average PRS (i.e., variable penetrance). Recent results in the UK Biobank49 have estimated that familial hypercholesterolemia carriers had an average cumulative CAD risk of 41% by age 75, but the risk varied between 20% to 80% depending on the PRS (GPSCAD in this case) for that individual. In addition, individuals with high PRS who were not familial hypercholesterolemia carriers had similar lifetime risk of CAD as carriers with low PRS.
These results indicate that for the vast majority of the population who do not carry CVD-related monogenic variants, the standard combined PRS and traditional risk factor score will be adequate. However, for monogenic carriers, having the PRS information can still be informative regarding their risk; importantly, the same PRS can be used regardless of carrier status. While similar rare variant analyses have not yet been performed for stroke, it is likely that the findings from other cardiovascular diseases will apply to stroke as well, and further work is needed to better understand how PRS can modify monogenic penetrance across a range of cardiovascular diseases and related traits.
Future Directions in PRS Development
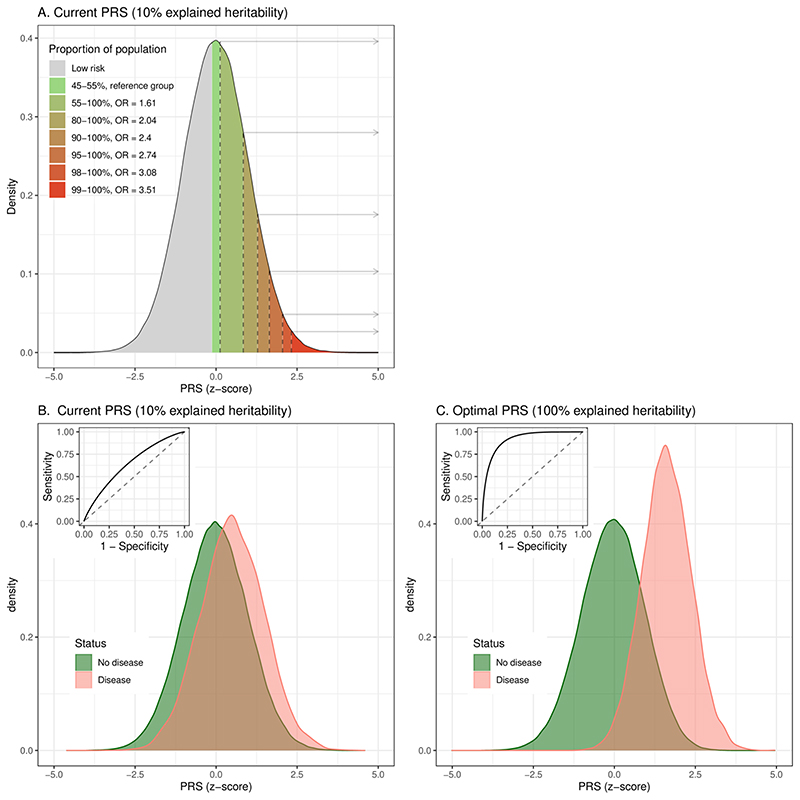
Several factors represent challenges for development of far more powerful and effective PRS for CVD. GWAS sample sizes will remain a critical factor across all PRS, since they directly affect the precision of the GWAS effect size estimates which, in turn, impact the PRS predictive power. As larger GWAS cohorts become available, new PRS will become more powerful, with the only limitation being the heritability of CVD50 (Figure 1). However, recruitment, genotyping, and phenotyping of large cohorts is costly and time-consuming. In the meantime, statistical approaches can be used to augment the effective sample size via the use of proxy designs (taking advantage of individuals with family history of CVD in addition to cases)51, methods that perform multi-trait GWAS52 to increase statistical power to detect CVD-associated loci, and PRS that combine multiple related PRS into a more powerful single score (metaGRS)31, 35.
Figure 1. Distribution of typical current and future PRS in the population.
(A) Stratification of risk for typical current CVD PRSs (explaining 10% of heritability), comparing the top bins in the populations versus the middle PRS category (45–55%) in terms of odds ratio (OR). (B) Distributions of PRS in disease and non-disease individuals for a typical current CVD PRS, as well as receiver-operating characteristic curves (ROC; AUC=0.645). (C) Discrimination for a hypothetical future PRS explaining all CVD heritability (AUC=0.914). Based on simulations of the liability threshold model50 assuming a CVD population prevalence of 3% and heritability of 50%.
Sex differences in CVD is another important topic for consideration in future PRS analyses. There are well-known differences in both incidence and presentation of CVD between the sexes, suggesting that sex-specific PRS may be better than sex-agnostic PRS. However, most GWAS are limited to the autosomal chromosomes. So far, no associations have been detected on chromosome X for CAD53. Yet, there may be some sex-specific effects for the same autosomal loci; these analyses would require performing a sex-specific GWAS of existing cohorts and may enable development of sex-specific PRS with better predictive power than existing PRS that do not consider sex-specific effects.
Another substantial challenge is ancestry, as it can have a large effect on the performance and interpretation of PRS54, 55. The majority of past cohorts collected for population research have been dominated by European ancestries and this has fed into the GWAS performed for CVD-related traits. The PRS developed from these GWAS tend to perform best in European ancestries, with reduced performance in other ancestries, particularly African. This is likely due to differences in allele frequencies and linkage disequilibrium between ancestries, but may also reflect some differences in environmental effects such as in lifestyle or diet (i.e., gene-by-environment interactions). In some cases, existing PRS can undergo some re-tuning in order to increase transferability56. However, in general, there remains a strong need for validation of existing scores on a wider range of ancestries, as well as larger non-European participation in GWAS cohorts which would allow derivation of ancestry-specific PRS57. Notably, the widely-used Pooled Cohort Equations only allow for White and Black categories, again highlighting the need for better incorporation of more diverse ancestries in clinical risk scores as well.
Challenges and Next Steps in Stroke PRS Development
In addition to the challenges faced by CVD PRS as a whole, stroke poses special challenges for development of effective PRS.
While all cardiovascular diseases are heterogeneous to some degree, this is an issue particularly with stroke. Besides the major distinction between ischaemic and haemorrhagic stroke, there are further clinical subtypes of stroke with distinct underlying aetiological pathways, such as large artery atherosclerotic stroke (LAS), cardioembolic stroke (CES), and small vessel stroke (SVS). Although there is some overlap in the genetic factors between stroke subtypes, heterogeneity in the influence of genetic variants on different stroke subtypes is the norm, not the exception11, 58. The majority of loci identified in the largest stroke GWAS to date (MEGASTROKE), were associated with multiple, but never all, stroke subtypes58. GWAS in stroke subtypes has been challenging due to the small sample sizes with confirmed diagnoses of these subtypes. One notable example is SVS, which causes up to a quarter of all ischaemic strokes. SVS is the most common stroke subtype caused by monogenic disease and has a strong association with family history59, however, up until recently only a small number of SVS risk loci have been identified in GWAS. Outside of the known monogenic loci, GWAS initially identified the role of the 16q24.2 locus in SVS risk60, with three more loci having been identified with varying degrees of certainty (2q33, 14q22, 12q24)59. Besides increases in sample size, leveraging related traits using multi-trait GWAS seems promising to increase statistical power to detect more SVS-associated loci61.
Despite these advances in stroke genetics, the implications for polygenic risk scores are not immediately clear. Currently, GWAS sample sizes with confirmed diagnoses of stroke subtypes remain low, limiting predictive power and necessitating the use of a generic ischaemic stroke PRS35 or all-stroke PRS27. However, there may be benefit in the future from developing subtype-specific PRS when larger sample sizes are available.
Clinical Applications and Implementation of PRS
A recent retrospective analysis of a stroke PRS examined five randomised controlled trials (ENGAGE AF-TIMI 48, SOLID-TIMI 52, SAVOR-TIMI 53, PEGASUS-TIMI 54, and FOURIER) totalling 51,000 participants (960 incident ischaemic stroke events)62. A 32-SNP score was more strongly associated with ischaemic stroke in participants without prior stroke than those with prior events, even after adjusting for a large number of clinical risk factors including hypertension, hyperlipidaemia, diabetes, smoking, atrial fibrillation, vascular diseases, and congestive heart failure (HR=1.24, 95% CI 1.05–1.45 for the top 33% of genetic risk vs the bottom 33% for participants without prior stroke). The score had similar predictive power across subgroups of sex and age. Further, in participants with atrial fibrillation and CHA2DS2-VASc of 2, being in the top 33% of genetic risk increased risk markedly (HR=3.97, 95% CI 1.04–15.2), with an absolute risk similar to that of a CHA2DS2-VASc of 3. These results further support the notion that stroke PRS are an independent risk factor outside of clinical risk factors, and that they may also be useful at identifying individuals with atrial fibrillation who are at increased stroke risk despite having moderate CHA2DS2-VASc scores of 2. Thus, stroke PRS may help guide anticoagulation therapy in individuals with atrial fibrillation. The use of a larger PRS with better predictive power35 would likely improve these results further and should be examined.
With the growing interest in PRS for CVD and other diseases, together with the wide range of cohorts and scores that have been developed, there is greater need for standardisation in terms of PRS curation and reporting. The recently-developed PGS Catalog63 serves as a central public repository for PRS for CVD, other diseases, and other human traits, where metadata regarding each score and its analysis are curated, including the performance metrics in each dataset, such as R2, area under the receiver-operating characteristic curve (AUC), and odds ratios. In addition, reporting standards for PRS have been recently developed (PRS Reporting Statement, PRS-RS)64, which call for publication of detailed information about each study population (recruitment method, demographics, exclusion criteria, definition of the clinical outcome) and the statistical analysis behind the development and validation of the score. The reporting standards aim to increase better transparency in PRS analysis, leading to higher reproducibility of results and ease of benchmarking of various competing scores.
Finally, we highlight two outstanding issues pertaining to deployment and communication of PRS to individuals. First, from a purely technical perspective, genotyping is essentially a one-time cost, and from the same set of genotypes (potentially augmented by statistical imputation of more variants) several PRS for CVD and other diseases can be calculated. The PRS can be updated later as new ones are developed. It remains to be seen how genotyping and scoring will be implemented within health systems to maintain consistent standards and protocols of care. A second important challenge is how to communicate PRS results to individuals in a consistent and transparent way that enables them, together with clinicians, to make the best informed decisions about CVD risk management. Preliminary results from the GeneRisk65 study in Finland indicate that interactive risk tools (such as their KardioKompassi tool) are effective in motivating high-risk individuals to adopt lifestyle changes, but more studies, including prospective66 and randomised controlled trials, are required to better understand the generalisability of these results and long-term impact of the risk information.
Conclusions
The past 15 years have seen immense development in genomics and genetics, enabling genome-wide association studies of large cohorts, and uncovering the ubiquitous polygenic basis for CVD, including stroke. The predictive power of polygenic scores for CVD now routinely surpasses that of traditional risk factors, and will likely increase in the future. The next generation of risk scores will likely integrate traditional risk factors together with polygenic risk, and also account for any monogenic variants a person may be carrying. Further research is needed to expand this approach to the various subtypes of stroke, better understand the role of sex in CVD genetics, better account for diverse ancestries in score development and validation, and finally, to safely and effectively communicate the risk results to individuals.
Acknowledgements
Sources of Funding
MI was supported by the Munz Chair of Cardiovascular Prediction and Prevention. This work was supported by the Victorian Government’s Operational Infrastructure Support (OIS) program, and by core funding from: the UK Medical Research Council (MR/L003120/1), the British Heart Foundation (RG/13/13/30194; RG/18/13/33946) and the National Institute for Health Research [Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust] [*]. This work was supported by Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), and the British Heart Foundation.
*The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Footnotes
Disclosures
GA reports speaker honoraria from Amgen One Cardiovascular Academy outside the submitted work. LRJ is a full-time employee of Hoffmann-La Roche Ltd. MI reports no conflicts of interest.
References
- 1.Meschia JF, Bushnell C, Boden-Albala B, Braun LT, Bravata DM, Chaturvedi S, Creager MA, Eckel RH, Elkind MS, Fornage M, et al. Guidelines for the primary prevention of stroke: a statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2014;45:3754–3832. doi: 10.1161/STR.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf PA, D’Agostino RB, Belanger AJ, Kannel WB. Probability of stroke: a risk profile from the Framingham Study. Stroke. 1991;22:312–318. doi: 10.1161/01.str.22.3.312. [DOI] [PubMed] [Google Scholar]
- 3.D’Agostino RB, Wolf PA, Belanger AJ, Kannel WB. Stroke risk profile: adjustment for antihypertensive medication. The Framingham Study. Stroke. 1994;25:40–43. doi: 10.1161/01.str.25.1.40. [DOI] [PubMed] [Google Scholar]
- 4.Goff DC, Jr, Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Gibbons R, Greenland P, Lackland DT, Levy D, O’Donnell CJ, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129:S49–73. doi: 10.1161/01.cir.0000437741.48606.98. [DOI] [PubMed] [Google Scholar]
- 5.Hippisley-Cox J, Coupland C, Brindle P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ. 2017;357:j2099. doi: 10.1136/bmj.j2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, Catapano AL, Cooney MT, Corra U, Cosyns B, Deaton C, et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts)Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR) Eur Heart J. 2016;37:2315–2381. doi: 10.1093/eurheartj/ehw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yusuf S, Joseph P, Rangarajan S, Islam S, Mente A, Hystad P, Brauer M, Kutty VR, Gupta R, Wielgosz A, et al. Modifiable risk factors, cardiovascular disease, and mortality in 155 722 individuals from 21 high-income, middle-income, and low-income countries (PURE): a prospective cohort study. Lancet. 2020;395:795–808. doi: 10.1016/S0140-6736(19)32008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khera AV, Won HH, Peloso GM, Lawson KS, Bartz TM, Deng X, van Leeuwen EM, Natarajan P, Emdin CA, Bick AG, et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J Am Coll Cardiol. 2016;67:2578–2589. doi: 10.1016/j.jacc.2016.03.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan RY, Markus HS. Monogenic causes of stroke: now and the future. J Neurol. 2015;262:2601–2616. doi: 10.1007/s00415-015-7794-4. [DOI] [PubMed] [Google Scholar]
- 10.Choi JC. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy: a genetic cause of cerebral small vessel disease. J Clin Neurol. 2010;6:1–9. doi: 10.3988/jcn.2010.6.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 14.van der Harst P, Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakker MK, van der Spek RAA, van Rheenen W, Morel S, Bourcier R, Hostettler IC, Alg VS, van Eijk KR, Koido M, Akiyama M, et al. Genome-wide association study of intracranial aneurysms identifies 17 risk loci and genetic overlap with clinical risk factors. Nature Genetics. 2020;52:1303–1313. doi: 10.1038/s41588-020-00725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, Iribarren C, Chakravarti A, Risch N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017;49:54–64. doi: 10.1038/ng.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, Ntalla I, Surendran P, Liu C, Cook JP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, Torstenson ES, Kovesdy CP, Sun YV, Wilson OD, et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019;51:51–62. doi: 10.1038/s41588-018-0303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPherson R, Tybjaerg-Hansen A. Genetics of Coronary Artery Disease. Circ Res. 2016;118:564–578. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Qi G, Park JH, Chatterjee N. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat Genet. 2018;50:1318–1326. doi: 10.1038/s41588-018-0193-x. [DOI] [PubMed] [Google Scholar]
- 22.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, Highland HM, Patel YM, Sorokin EP, Avery CL, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield MJ, Devlin JJ, Nordio F, Hyde CL, Cannon CP, Sacks FM, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tada H, Shiffman D, Smith JG, Sjogren M, Lubitz SA, Ellinor PT, Louie JZ, Catanese JJ, Engstrom G, Devlin JJ, et al. Twelve-single nucleotide polymorphism genetic risk score identifies individuals at increased risk for future atrial fibrillation and stroke. Stroke. 2014;45:2856–2862. doi: 10.1161/STROKEAHA.114.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tada H, Melander O, Louie JZ, Catanese JJ, Rowland CM, Devlin JJ, Kathiresan S, Shiffman D. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur Heart J. 2016;37:561–567. doi: 10.1093/eurheartj/ehv462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tikkanen E, Havulinna AS, Palotie A, Salomaa V, Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:2261–2266. doi: 10.1161/ATVBAHA.112.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MEGASTROKE Consortium, International Stroke Genetics Consortium. Rutten-Jacobs L, Larsson SC, Malik R, Rannikmae K, Sudlow C, Dichgans M, Markus HS, Traylor M. Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: follow-up of 306,473 UK Biobank participants. BMJ. 2018;363:k4168. doi: 10.1136/bmj.k4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abraham G, Havulinna AS, Bhalala OG, Byars SG, De Livera AM, Yetukuri L, Tikkanen E, Perola M, Schunkert H, Sijbrands EJ, et al. Genomic prediction of coronary heart disease. Eur Heart J. 2016;37:3267–3278. doi: 10.1093/eurheartj/ehw450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik R, Bevan S, Nalls MA, Holliday EG, Devan WJ, Cheng YC, Ibrahim-Verbaas CA, Verhaaren BF, Bis JC, Joon AY, et al. Multilocus genetic risk score associates with ischemic stroke in case-control and prospective cohort studies. Stroke. 2014;45:394–402. doi: 10.1161/STROKEAHA.113.002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubitz SA, Yin X, Lin HJ, Kolek M, Smith JG, Trompet S, Rienstra M, Rost NS, Teixeira PL, Almgren P, et al. Genetic Risk Prediction of Atrial Fibrillation. Circulation. 2017;135:1311–1320. doi: 10.1161/CIRCULATIONAHA.116.024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, Lai FY, Kaptoge S, Brozynska M, Wang T, et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J Am Coll Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mars N, Koskela JT, Ripatti P, Kiiskinen TTJ, Havulinna AS, Lindbohm JV, Ahola-Olli A, Kurki M, Karjalainen J, Palta P, et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med. 2020;26:549–557. doi: 10.1038/s41591-020-0800-0. [DOI] [PubMed] [Google Scholar]
- 34.Weng LC, Preis SR, Hulme OL, Larson MG, Choi SH, Wang B, Trinquart L, McManus DD, Staerk L, Lin H, et al. Genetic Predisposition, Clinical Risk Factor Burden, and Lifetime Risk of Atrial Fibrillation. Circulation. 2018;137:1027–1038. doi: 10.1161/CIRCULATIONAHA.117.031431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham G, Malik R, Yonova-Doing E, Salim A, Wang T, Danesh J, Butterworth AS, Howson JMM, Inouye M, Dichgans M. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat Commun. 2019;10:5819. doi: 10.1038/s41467-019-13848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hachiya T, Hata J, Hirakawa Y, Yoshida D, Furuta Y, Kitazono T, Shimizu A, Ninomiya T. Genome-Wide Polygenic Score and the Risk of Ischemic Stroke in a Prospective Cohort: The Hisayama Study. Stroke. 2020;51:759–765. doi: 10.1161/STROKEAHA.119.027520. [DOI] [PubMed] [Google Scholar]
- 39.Wunnemann F, Sin Lo K, Langford-Avelar A, Busseuil D, Dube MP, Tardif JC, Lettre G. Validation of Genome-Wide Polygenic Risk Scores for Coronary Artery Disease in French Canadians. Circ Genom Precis Med. 2019;12:e002481. doi: 10.1161/CIRCGEN.119.002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dikilitas O, Schaid DJ, Kosel ML, Carroll RJ, Chute CG, Denny JA, Fedotov A, Feng Q, Hakonarson H, Jarvik GP, et al. Predictive Utility of Polygenic Risk Scores for Coronary Heart Disease in Three Major Racial and Ethnic Groups. Am J Hum Genet. 2020;106:707–716. doi: 10.1016/j.ajhg.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S, Genovese G, Loh PR, Bhatia G, Do R, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Regeneron Genetics C. Hindy G, Aragam KG, Ng K, Chaffin M, Lotta LA, Baras A, Drake I, Orho-Melander M, Melander O, et al. Genome-Wide Polygenic Score, Clinical Risk Factors, and Long-Term Trajectories of Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2020;40:2738–2746. doi: 10.1161/ATVBAHA.120.314856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pencina MJ, Navar AM, Wojdyla D, Sanchez RJ, Khan I, Elassal J, D’Agostino RB, Sr, Peterson ED, Sniderman AD. Quantifying Importance of Major Risk Factors for Coronary Heart Disease. Circulation. 2019;139:1603–1611. doi: 10.1161/CIRCULATIONAHA.117.031855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331–344. doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Widen E, Surakka I, Mars N, Pollanen P, Hotakainen K, Partanen J, Aro J, Ripatti S. Returning cardiovascular disease risk prediction back to individuals motivate beneficial lifestyle changes: Preliminary results from the GeneRISK-study; 51st European Society of Human Genetics Conference; 2018. [Google Scholar]
- 46.Sun L, Pennells L, Kaptoge S, Nelson CP, Ritchie SC, Abraham G, Arnold M, Bell S, Bolton T, Burgess S, et al. Polygenic risk scores in cardiovascular risk prediction: A cohort study and modelling analyses. PLo SMed. 2021;18:e1003498. doi: 10.1371/journal.pmed.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hynninen Y, Linna M, Vilkkumaa E. Value of genetic testing in the prevention of coronary heart disease events. PLoS One. 2019;14:e0210010. doi: 10.1371/journal.pone.0210010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navar-Boggan AM, Peterson ED, D’Agostino RB, Sr, Neely B, Sniderman AD, Pencina MJ. Hyperlipidemia in early adulthood increases long-term risk of coronary heart disease. Circulation. 2015;131:451–458. doi: 10.1161/CIRCULATIONAHA.114.012477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fahed AC, Wang M, Homburger JR, Patel AP, Bick AG, Neben CL, Lai C, Brockman D, Philippakis A, Ellinor PT, et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun. 2020;11:3635. doi: 10.1038/s41467-020-17374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu JZ, Erlich Y, Pickrell JK. Case-control association mapping by proxy using family history of disease. Nat Genet. 2017;49:325–331. doi: 10.1038/ng.3766. [DOI] [PubMed] [Google Scholar]
- 52.Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, Nguyen-Viet TA, Wedow R, Zacher M, Furlotte NA, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loley C, Alver M, Assimes TL, Bjonnes A, Goel A, Gustafsson S, Hernesniemi J, Hopewell JC, Kanoni S, Kleber ME, et al. No Association of Coronary Artery Disease with X-Chromosomal Variants in Comprehensive International Meta-Analysis. Sci Rep. 2016;6:35278. doi: 10.1038/srep35278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang M, Menon R, Mishra S, Patel AP, Chaffin M, Tanneeru D, Deshmukh M, Mathew O, Apte S, Devanboo CS, et al. Validation of a Genome-Wide Polygenic Score for Coronary Artery Disease in South Asians. J Am Coll Cardiol. 2020;76:703–714. doi: 10.1016/j.jacc.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakaue S, Kanai M, Karjalainen J, Akiyama M, Kurki M, Matoba N, Takahashi A, Hirata M, Kubo M, Matsuda K, et al. Trans-biobank analysis with 676,000 individuals elucidates the association of polygenic risk scores of complex traits with human lifespan. Nat Med. 2020;26:542–548. doi: 10.1038/s41591-020-0785-8. [DOI] [PubMed] [Google Scholar]
- 58.Traylor M, Anderson CD, Rutten-Jacobs LCA, Falcone GJ, Comeau ME, Ay H, Sudlow CLM, Xu H, Mitchell BD, Cole JW, et al. Subtype Specificity of Genetic Loci Associated With Stroke in 16 664 Cases and 32 792 Controls. Circ Genom Precis Med. 2019;12:e002338. doi: 10.1161/CIRCGEN.118.002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marini S, Anderson CD, Rosand J. Genetics of Cerebral Small Vessel Disease. Stroke. 2020;51:12–20. doi: 10.1161/STROKEAHA.119.024151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Traylor M, Malik R, Nalls MA, Cotlarciuc I, Radmanesh F, Thorleifsson G, Hanscombe KB, Langefeld C, Saleheen D, Rost NS, et al. Genetic variation at 16q24.2 is associated with small vessel stroke. Ann Neurol. 2017;81:383–394. doi: 10.1002/ana.24840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung J, Marini S, Pera J, Norrving B, Jimenez-Conde J, Roquer J, Fernandez-Cadenas I, Tirschwell DL, Selim M, Brown DL, et al. Genome-wide association study of cerebral small vessel disease reveals established and novel loci. Brain. 2019;142:3176–3189. doi: 10.1093/brain/awz233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marston NA, Patel PN, Kamanu FK, Nordio F, Melloni GM, Roselli C, Gurmu Y, Weng LC, Bonaca MP, Giugliano RP, et al. Clinical Application of a Novel Genetic Risk Score for Ischemic Stroke in Patients With Cardiometabolic Disease. Circulation. 2021;143:470–478. doi: 10.1161/CIRCULATIONAHA.120.051927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambert SA, Gil L, Jupp S, Ritchie SC, Xu Y, Buniello A, McMahon A, Abraham G, Chapman M, Parkinson H, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat Genet. 2021 doi: 10.1038/s41588-021-00783-5. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wand H, Lambert SA, Tamburro C, Iacocca MA, O’Sullivan JW, Sillari C, Kullo IJ, Rowley R, Dron JS, Brockman D, et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature. 2021;591:211–219. doi: 10.1038/s41586-021-03243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Widen E, Junna N, Ruotsalainen S, Surakka I, Mars N, Ripatti P, Partanen JJ, Aro J, Mustonen P, Tuomi T, et al. Communicating polygenic and non-genetic risk for atherosclerotic cardiovascular disease - An observational follow-up study. medRxiv. 2020:2020.2009.2018.20197137. doi: 10.1161/CIRCGEN.121.003459. [DOI] [PubMed] [Google Scholar]
- 66.Yanes T, Meiser B, Young MA, Kaur R, Mitchell G, Barlow-Stewart K, Roscioli T, Halliday J, James P. Psychosocial and behavioral impact of breast cancer risk assessed by testing for common risk variants: protocol of a prospective study. BMC Cancer. 2017;17:491. doi: 10.1186/s12885-017-3485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]