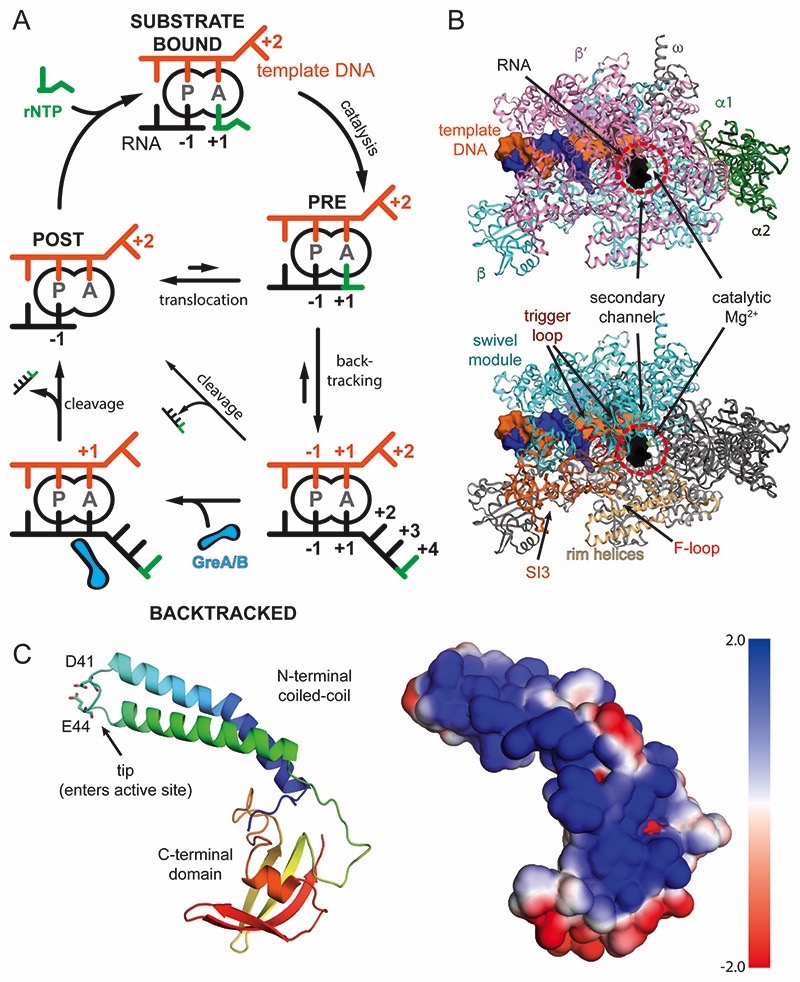
Figure 1. Schematic of transcription, RNAP, and GreB.
(A) Nucleotide addition and backtracking: In the post-translocated state (POST), the RNA 3′ end occupies the P-site, while the A-site holds the +1 template DNA, which dictates the next rNTP substrate to bind (green, top). Catalysis extends the RNA giving rise to the pre-translocated state (PRE). RNAP translocates relative to DNA by one base pair and concludes the cycle. Misincorporations, weak RNA-DNA hybrids or pause signals can cause backtracking and extrusion of the RNA from the active site (bottom, right). Reactivation requires forward translocation (slow, upward arrow), intrinsic RNA cleavage (slow, diagonal arrow), or Gre-factor assisted cleavage (bottom, left). (B) RNAP structure: View into the secondary channel with the five subunits, DNA, and RNA indicated (top). RNAP modules, which will be referred to throughout the manuscript are indicated (bottom) (C) GreB contains two domains. The N-terminal coiled-coil inserts its tip with two conserved acidic residues into the active site through the secondary channel. The C-terminal domain interacts with the RNAP surface. The electrostatic surface potential of GreB shows one side is highly positively charged and predicted to interact with backtracked RNA (right).