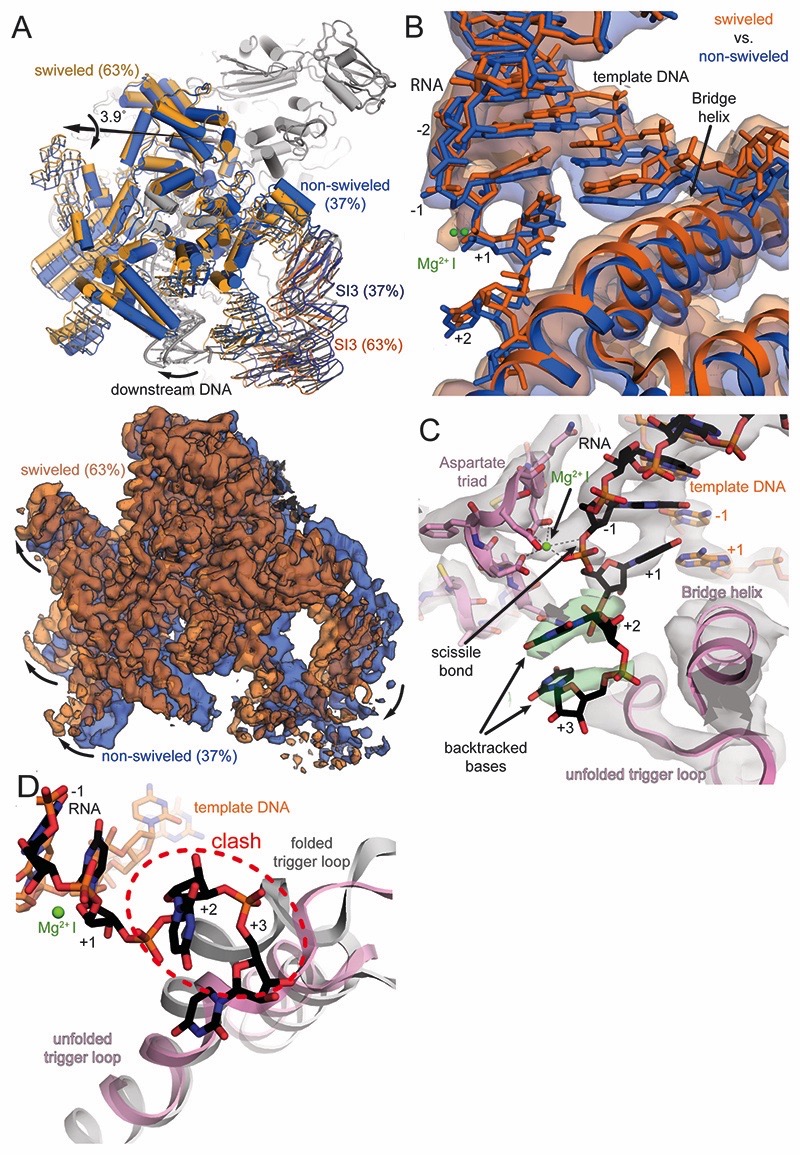
Figure 2. RNAP and active site conformation of backtracked complex.
(A) The swivel module (clamp and shelf domain of RNAP, shades of blue and orange) can rotate relative to the core module by almost 4˚ in the backtracked complex. The trigger loop insertion domain (SI3, orange and blue) and downstream DNA move along with the swivel module. Superposition of EM maps from the two 3D classes (blue 37%, orange 63%) confirms swiveling. (B) Downstream DNA movement affects the downstream end of the RNA-DNA hybrid. The template DNA shifts around 2Å between the two conformations and affects the RNA in A- (+1) and P-site (-1). The two positions are reminiscent of an oscillation around the position observed in an EC (Kang et al., 2017). (C) Cryo-EM density (grey and green transparent surface) for the active site reveals the backtracked RNA at lower contour level (black sticks and green surface), the BH, unfolded TL, and Aspartate triad (pink), the template DNA (orange) and MgI (green). (D) Superposition of a substrate bound EC shows the steric clash between the folded trigger loop (grey) with the backtracked RNA (black) in the present reconstruction.