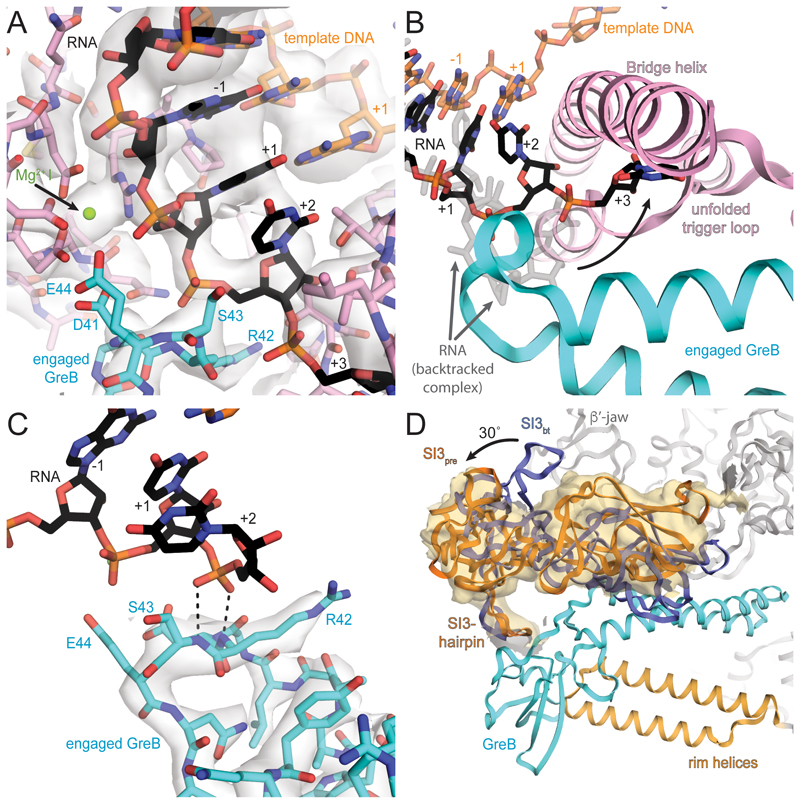
Figure 3. Active site of pre-cleavage complex and GreB interaction with RNAP.
(A) Cryo-EM density (light grey, transparent surface) for the active site reveals density for the RNA (black) and template DNA (orange), the Aspartate triad and MgI (pink and green respectively) and GreB (cyan). (B) Comparing the backtracked complex (RNA transparent grey) with the pre-cleavage complex (RNA black), shows GreB shifts the backtracked portion of the RNA towards the BH and unfolded TL (pink). (C) A different view of the active site shows well-resolved density (grey transparent surface) for the tip of GreB (cyan). The backtracked RNA base in position +2 (black), stabilizes the backbone of GreB and helps to orient the catalytic residues. (D) GreB (cyan) interacts through the CTD with the secondary channel rim helices (light orange) and the trigger loop insertion SI3 (orange, SI3pre), which rotates almost 30° relative to the orientation in the backtracked state (blue, SI3bt). A low-pass filtered map confirms SI3 rotation (orange surface).