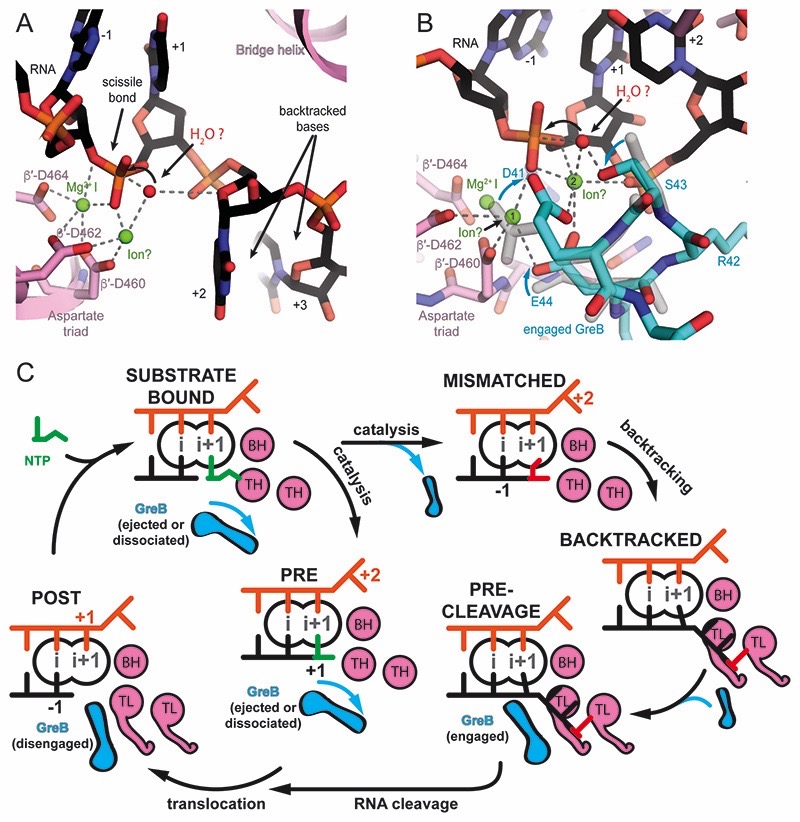
Figure 6. Models for RNA cleavage and the role of GreB during transcription elongation.
(A) In the backtracked complex, the nucleophile (red sphere) can be placed in line with the scissile bond. The O4′ of the first backtracked base (+2, black) may help to coordinate the nucleophile. MgII (modelled), could be coordinated by β′-D460, and β′-D462, but would be 3Å away from the attacking nucleophile. Small changes in the RNA backbone conformation may allow to adopt optimal geometry for the nucleophilic attack. (B) In the pre-cleavage complex, the nucleophile (red sphere) can be placed in line with the scissile bond. MgII can be modeled to be coordinated by β′-D460, β′-D462, the phosphate of the base in the A-site (+1) as well as GreB D41 and E44. However, this position (position 1) is 5Å away from the nucleophile. A change in the RNA backbone conformation may allow direct coordination of the nucleophile by MgII. Alternatively, a hypothetical third ion (modelled in position 2) as observed in DNA polymerase could activate the nucleophile. S43 in GreB might help orient the nucleophile consistent with a subtle decrease in cleavage rates upon mutation to Alanine. (C) Model for GreB’s role in transcription elongation: During a canonical elongation cycle (left), GreB cannot access the active site of substrate-bound and pre-translocated complexes because of the folded TH and secondary channel closure (not shown). As a result of erroneous incorporations or pause inducing DNA sequences, RNAP can backtrack and extrude the RNA 3′-end from the active site (right). RNA backtracking by 2 or more bases results in an unfolded TL and allows GreB to access the active site. GreB accelerates RNA cleavage and gives rise to a post-translocated EC, which can resume transcription (bottom).