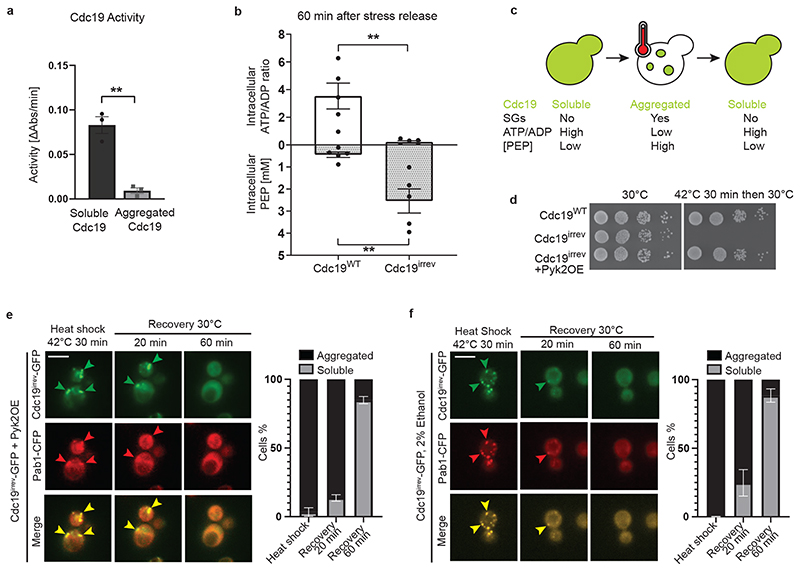
Figure 2. SG disassembly depends on Cdc19 re-solubilization and restoration of ATP production.
(A) The activity of purified Cdc19WT before (soluble) or after a 30 min heat shock at 42 °C (aggregated) was measured using a lactate dehydrogenase-coupled activity assay and plotted as the decrease in absorbance at 340 nm per min. Mean ± S.E.M. of n = 3 independent experiments is indicated (two-tailed t-test, P = 0.0018).
(B) cdc19irrev cells are unable to re-activate Cdc19 and resume ATP production after stress release. Intracellular ATP/ADP ratio and phosphoenolpyruvate (PEP) concentrations (mM) were determined in wild-type and cdc19irrev cells, after a 30 min heat shock at 42 °C and recovery at 30 °C for 60 min. Mean ± S.E.M. of n = 5 independent experiments is shown (two-tailed Mann-Whitney test, PATP/ADP = 0.0079, PPEP = 0.0079).
(C) Schematic drawing showing a yeast cell before, during, and after heat stress. Upon stress, Cdc19 (represented in green) forms inactive aggregates, which colocalize with SGs. Energy levels (ATP/ADP ratio) rapidly decrease under these conditions, while Cdc19’s substrate phosphoenolpyruvate (PEP) accumulates. Upon stress release, Cdc19 re-solubilizes, gets re-activated, and consumes PEP to produce ATP, concomitant with SG disassembly.
(D) Serial dilutions of exponentially growing cells of the indicated genotype were spotted on agar plates before or after a 30 min heat shock at 42 °C, and imaged after 3 days at 30 °C (n = 3 independent experiments).
(E) – (F) ATP production using Cdc19-independent pathways allows Cdc19irrev aggregate and SG re-solubilization after stress release. Cells of the indicated genotype were grown at 30 °C with 2% glucose (E) or 2% ethanol (F) as carbon source and subjected to heat stress for 30 min at 42 °C before recovery at 30 °C in the presence of 25 μg/ml cycloheximide to prevent de novo protein synthesis. Representative images are shown as in Fig. 1A. Plots indicate mean percentage (%) of cells with Cdc19 aggregates ± S.E.M (n = 3 independent experiments, >30 cells per sample per experiment). Scale bar: 5 μm. Source data for all graphical representations can be found in Source Data Fig. 2.