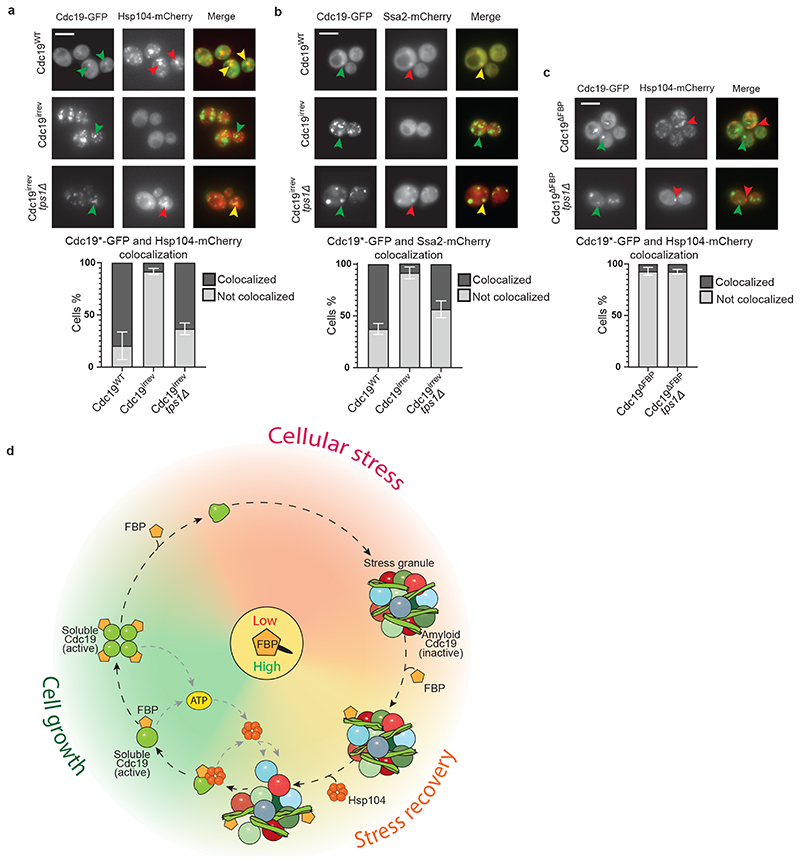
Figure 6. FBP binding to Cdc19 amyloids promotes chaperone recruitment triggering Cdc19 re-solubilization and SG disassembly in vivo .
(A) – (C) FBP binding to Cdc19 amyloids promotes chaperone recruitment, which correlates with aggregate disassembly. Wild-type or tps1Δ cells co-expressing mCherry-tagged Hsp104 (A, C) or Ssa2 (B) and either GFP-tagged Cdc19WT, Cdc19irrev or Cdc19ΔFBP were subjected to heat stress for 30 min at 42 °C and imaged by fluorescence microscopy. cdc19ΔFBP cells (C) overexpress Pyk2 to restore ATP production and cell viability. Representative GFP- and mCherry-images are shown, together with the merged image to visualize co-localization of Cdc19 aggregates and the chaperones (n = 3 independent experiments). Scale bar: 5 μm. Plots indicate mean percentage (%) of cells with co-localized Cdc19 and Hsp104 or Ssa2 foci ± S.E.M (n = 3 independent experiments, >300 cells per sample per experiment).
(D) Molecular network coupling cellular metabolism with reversible Cdc19 amyloids and stress granule dynamics. During cell growth, the metabolite FBP is produced through glycolysis and its binding to Cdc19 promotes the formation of active, non-aggregation-prone Cdc19 tetramers. Upon stress, FBP levels rapidly drop, leading to tetramer disassembly and the accumulation of Cdc19 monomers. In this state, Cdc19 exposes its aggregation-prone LCR and is primed to form Cdc19 amyloids, which are inactive and accumulate in SGs. After stress release, FBP levels rise and FBP directly binds to Cdc19 amyloids, thereby facilitating the recruitment of Hsp104 and Ssa2 to the aggregates. These chaperones allow re-solubilisation of Cdc19 amyloids and the release of Cdc19 monomers. Re-solubilised, active Cdc19 in turn uses PEP to produce ATP, which is required to activate chaperones to further solubilize Cdc19 and increase energy production. This positive feedback loop fuels disassembly of Cdc19 amyloids and other SG components. Concomitant breakdown of trehalose liberates glucose moieties for glycolysis and modulates cytoplasmic diffusion. Orchestrated disassembly of SGs then releases several mRNAs and metabolic enzymes, and allows full resumption of cell growth and cell cycle progression. Source data for all graphical representations can be found in Source Data Fig. 6.