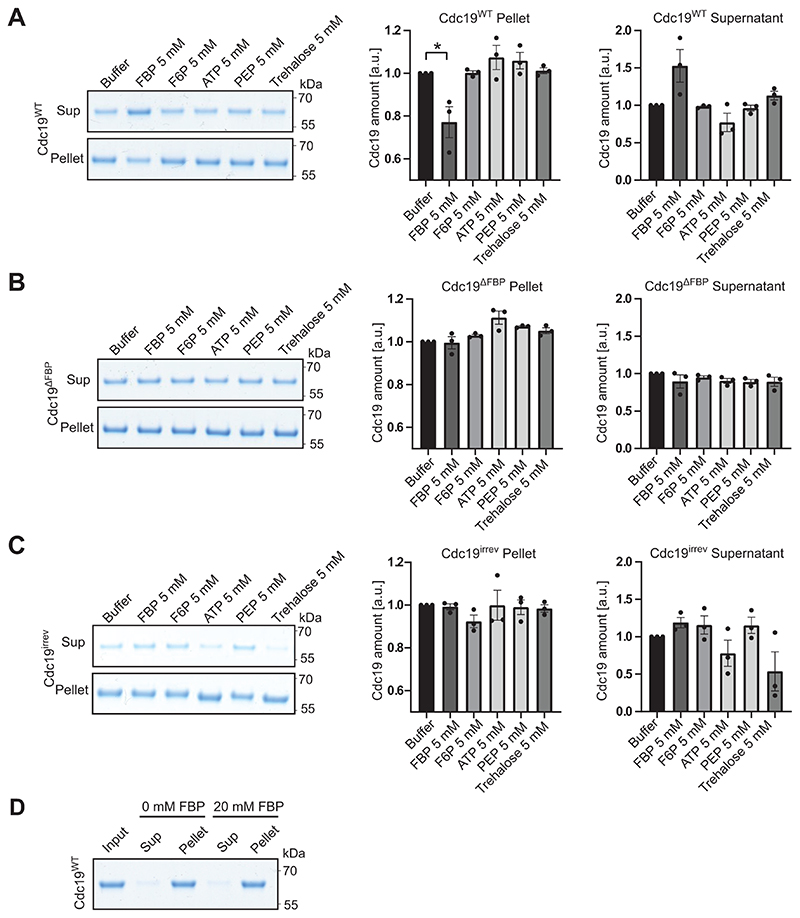
Extended Data Fig. 3. FBP specifically reduces aggregation of purified Cdc19WT, but not Cdc19ΔFBP and Cdc19irrev mutant proteins.
(A)– (C) Purified wild-type Cdc19 (A), FBP binding-deficient Cdc19ΔFBP mutant (B) or Cdc19irrev mutant (C) proteins were mixed as indicated with 5 mM FBP or for control 5 mM F6P, 5 mM ATP, 5 mM PEP, 5 mM trehalose or buffer and incubated at 30 °C for 14 hours. Cdc19 aggregates were separated from soluble protein by centrifugation and the supernatant (Sup) and pellet (Pellet) fractions were analysed by SDS-PAGE and Coomassie blue staining. Cdc19 amount was quantified in the Pellet and Supernatant by measuring Cdc19 band intensities using ImageJ and normalizing to buffer controls, and is displayed as mean and S.E.M. of three independent experiments (two-tailed t-test, P = 0.0342).
(D) Addition of FBP alone is not sufficient to re-solubilize pre-formed Cdc19WT amyloids in vitro. Purified Cdc19WT (Input) was incubated for 10 min at 42 °C to trigger its aggregation, and the resulting amyloids were incubated with (20 mM) or without (0 mM) FBP for several hours. Cdc19 re-solubilization was then tested by centrifugation and analysis of the resulting supernatant (Sup) and pellet (Pellet) fractions by SDS-PAGE and Coomassie blue staining. The image is representative of three independent experiments. Unprocessed original scans of gels are shown in Source Data Extended Data Fig. 3.