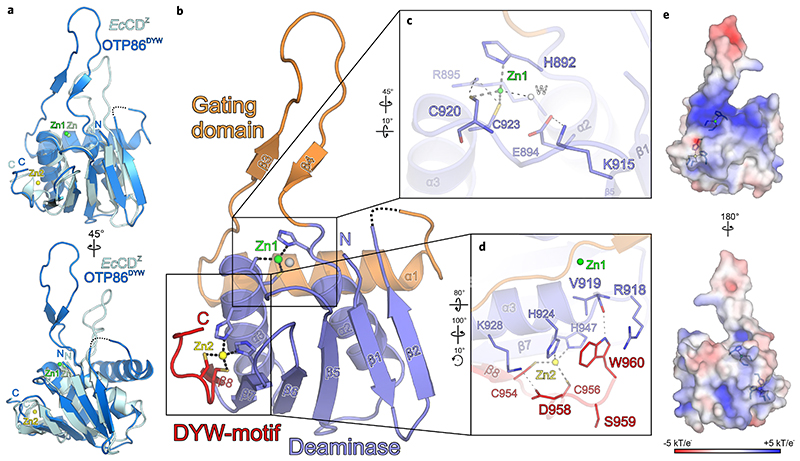
Fig. 1. Crystal structure of the A. thaliana OTP86 DYW domain.
a, Superimposition of OTP86DYW (marine) with E. coli cytidine deaminase (EcCDZ, cyan) bound to the inhibitor zebularine (not shown) (PDB-ID: 1CTU; ref. 32). The consensus deaminase zinc ions are shown as green (OTP86DYW, Zn1) and light-green (EcCDZ, Zn) spheres, a zinc ion partially coordinated by the DYW motif is shown in yellow (Zn2). b, The OTP86DYW structure defines a paradigmatic organization for DYW domains. The cytidine deaminase domain (slate) coordinates a zinc ion (Zn1, green) three-fold with H892, C920 and C923, the fourth position is occupied by a water molecule (W, white sphere). The deaminase domain is interrupted by a gating domain (orange) and terminates with a DYW motif (red), partially coordinating a second zinc ion (Zn2, yellow). c, A close-up view on the cytidine deaminase active site, with catalytically relevant residues shown as sticks. d, A close-up view of the DYW motif and the flanking β-strand 7 as well as α-helix 3. e, Electrostatic surface potentials as indicated by the colour scale bar (bottom), obtained by APBS version 1.5 and plotted on the surface of OTP86DYW. Residues involved in zinc coordination are shown as sticks; zinc atoms are as in b. Rotation symbols indicate the views relative to b. Interacting residues are shown as sticks and coloured by atom type. Blue, nitrogen; red, oxygen; yellow, sulfur; carbons take the colour of the respective molecule. Dashed lines represent hydrogen bonds, whereas thick grey dashed lines indicate zinc coordination. Dashed lines in the ribbon plots represent residues 842-844 not clearly defined by electron density.