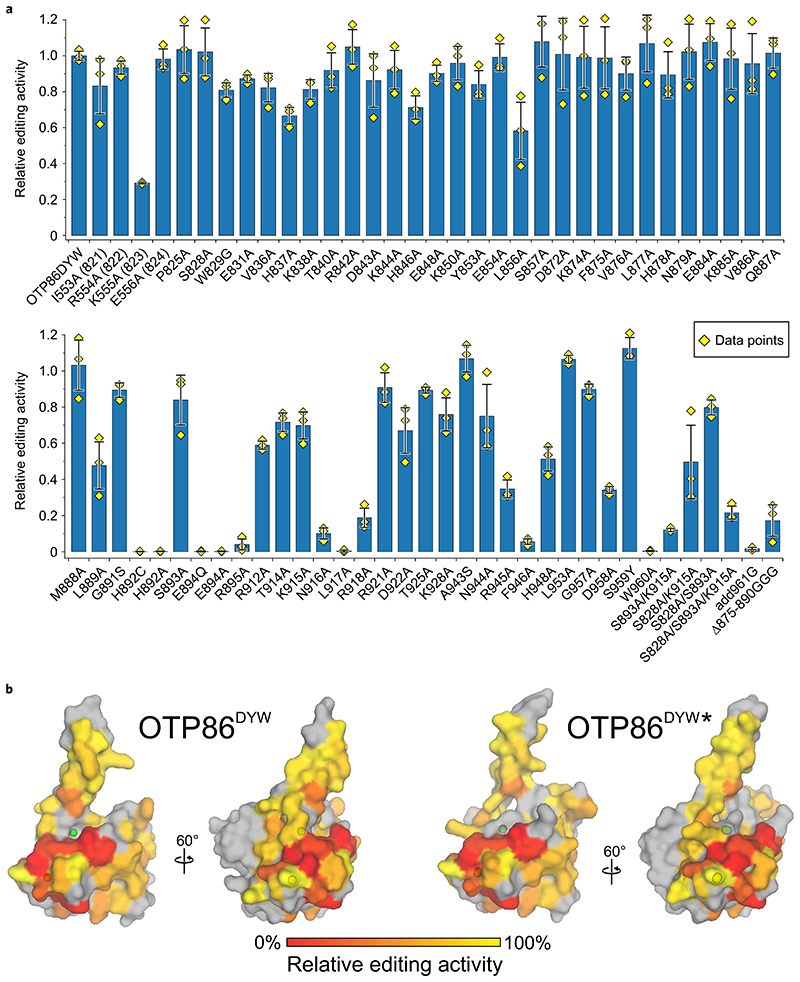
Fig. 5. Orthogonal in vivo RNA editing validates the OTP86DYW domain structure and activation.
a, The C-to-U editing activities at the nad4eU272SL site in E. coli expressed with the PPR56PPRE1E2-OTP86DYW fusion protein (OTP86DYW), or its mutants, are plotted. The activities of mutants are relative to that of PPR56PPRE1E2-OTP86DYW (82.4 ± 2.1% edited). The bars represent the mean values, with each mutant protein ±s.d. based on three independent experiments (shown as yellow diamonds). The soluble protein expression of each mutated construct in E. coli was verified by western blot analysis (Supplementary Fig. 7). b, The activities of OTP86DYW mutants shown in a plotted on the surface of the inactive OTP86DYW and the activated OTP86DYW★ structure as a heatmap (activity is scaled in the bar on the bottom), with untested residues shown in grey.