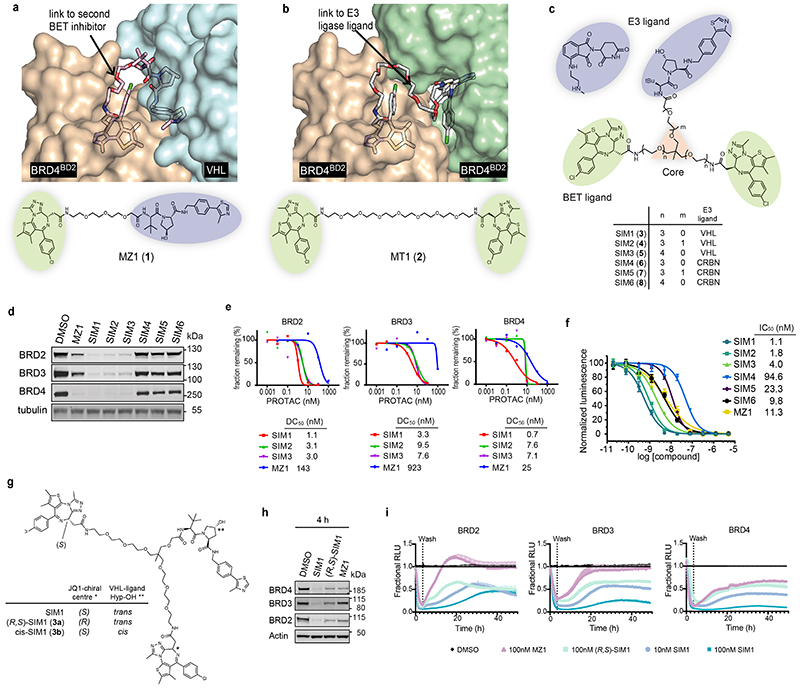
Figure 1. Structure-inspired design of trivalent PROTACs identifies VHL-based SIM1 as the most potent BET degrader.
a,b) Inspection of ternary complex crystal structures of VHL:MZ1:BRD4BD2 (a, PDB:5T35) and BRD4BD2:MT1:BRD4BD2 (b, PDB 5JWM) guided the identification of solvent-exposed region for chemical branching of linkers in trivalent PROTAC design. Chemical structures of parent bivalent molecules MZ1 and MT1 are shown. c) Chemical structures of designed trivalent PROTACs SIM1-6 based on VHL and CRBN E3 ligase ligands. d) Immunoblot analysis of BRD2, BRD3, BRD4 after treatment of HEK293 cells with 1μM PROTACs or DMSO for 4h, performed as n=1. Full blots are supplied as Source Data Fig. 1. e) Protein levels of BRD2, BRD3, BRD4 in HEK293 cells treated with serially diluted PROTACs SIM1-SIM3 for 4h. Quantification of BET protein levels was done relative to DMSO control and shown plots used to measure the tabulated DC50 values. Corresponding blots are in Extended Data Fig. 1c, and full blots are supplied as Source Data Fig. 1. f) Cell viability of MV4;11 AML cell line following treatment with PROTACs or DMSO for 48h in three replicates for each concentration point. g) Chemical structures of SIM1 and its designed negative controls, (R,S)-SIM1 and cis-SIM1. Reversed stereocenters are indicated by asterisks. h) Immunoblot of degradation of BET proteins in HEK293 cells after treatment with indicated compounds at 1μM or DMSO for 4h. Full blots are supplied as Source Data Fig. 1. i) CRISPR HiBiT-BRD2, BRD3, and BRD4 HEK293 cells were treated with 100nM of DMSO, MZ1, (R,S)-SIM1, and both 10nM and 100nM of SIM1 in replicate plates for washout experiments. Media containing the 10nM and/or 100nM compounds was removed at 3.5h, indicated on the graphs, and replaced with media lacking compounds for the remainder of the experiment. Luminescence (RLU) was continuously monitored over a 50h time period and is plotted normalized to the DMSO control as Fractional RLU.