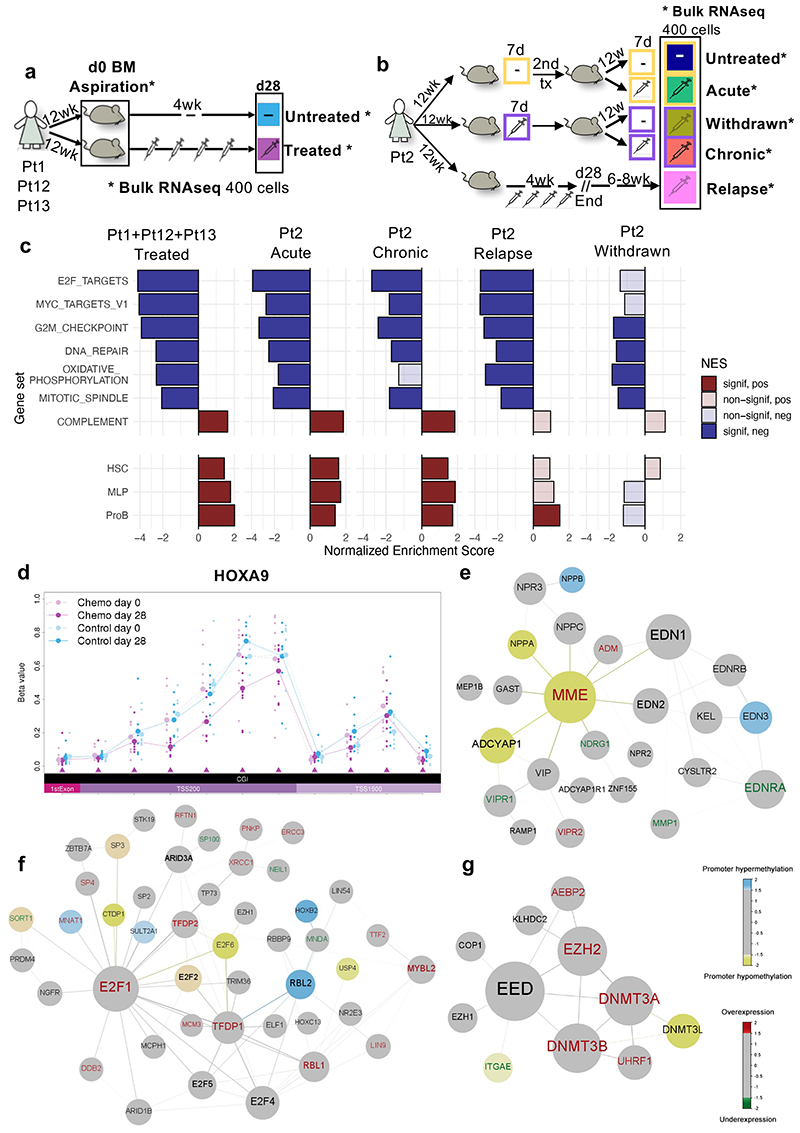
Figure 5. Transcriptionally driven phenotypes contribute to treatment resistance in BCP-ALL and DNA methylation analysis identifies epigenetically deregulated hotspots in BCP-ALL resistant cells.
(a-b) Graphical representation of the standard and multi-arm treatment models. In the standard model (a) matching d0 and d28 cells harvested from 9 untreated and 9 treated mice were analyzed. Untreated samples, shown in light blue, represent d0 cells from untreated mice and treated mice, as well as d28 samples from untreated mice. In the multi-arm model (b), two consecutive rounds of transplantation and treatment (7 days) allowed the analysis of ‘acutely treated’ (green), ‘treatment withdrawn’ (dark yellow), ‘chronically treated’ (red), and ‘untreated control’ (dark blue) cells harvested from 3 mice each (see main text). ‘Relapse-like’ cells were also obtained through a separate arm of the experiment, whereby 2 mice were treated for 28 days and then allowed to relapse in situ in the absence of further treatment (6-8 weeks). (c) Hallmark gene-sets and published signatures associated with predefined differentiation stages of the B-cell ontogeny32,47, which represent the core processes deregulated during chemotherapy across different patient subtypes and treatment regimens (see Supplementary Table 4 for the full list). Each population is compared to the corresponding untreated control. P-values are obtained using 2-sided permutation test48 with 10,000 permutations; significance thresholds: <0.05 after Benjamini-Hochberg multiple test correction. (d) Representative example of a region (HOXA9 locus) differentiallymethylated in d28 treated mice as compared to both d0 treated-mice and untreated-mice at d0/d28. All annotated CpG regions for the loci are listed at the bottom of the plot in chromosomal order. Each line shows the mean beta value of the corresponding sample. TSS200 = 0–200 bases upstream of the transcriptional start site (TSS). TSS1500 = 200–1500 bases upstream of the TSS. (e-f-g) Representative results of 3 out of 5 gene modules identified using the Functional Epigenetic Modules (FEM) algorithm31 comparing treated versus untreated d28 cells using the default FEM protein-protein interaction network. In each representation, the largest circle represents the ‘seeds’ (or master regulator) of the corresponding functional epigenetic module. In the figure, blue nodes represent significantly hypermethylated gene promoters, yellow nodes represent significantly hypomethylated gene promoters, red labels represent significantly overexpressed genes and green labels represent significantly underexpressed genes.