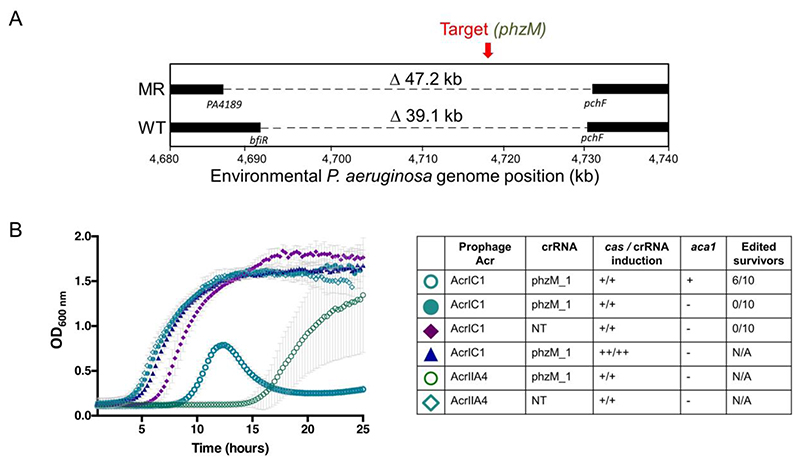
Figure 5.
A) Schematic of whole genome sequencing of an environmental isolate of PAO1 with an endogenous Type I-C system. Two survivors were isolated post-targeting using either wild-type (WT) direct repeats flanking the spacer, or modified repeats (MR). Bars indicate boundaries of deletions with ORF indicated below. B) Growth curves of PAO1IC lysogenized by recombinant DMS3m phage expressing acrIIA4 or acrIC1 from the native acr locus. CRISPR-Cas3 activity is induced with either 0.5mM (+) or 5mM (++) IPTG and 0.1% (+) or 0.3% (++) arabinose. Edited survivors reflect number of isolated survivor colonies missing the targeted gene (phzM) and ‘N/A’ means that editing was not assessed as no growth defect was seen. ‘NT’ means a non-targeting crRNA was expressed. Each growth curve is the average of 10 biological replicates and error bars represent SD.