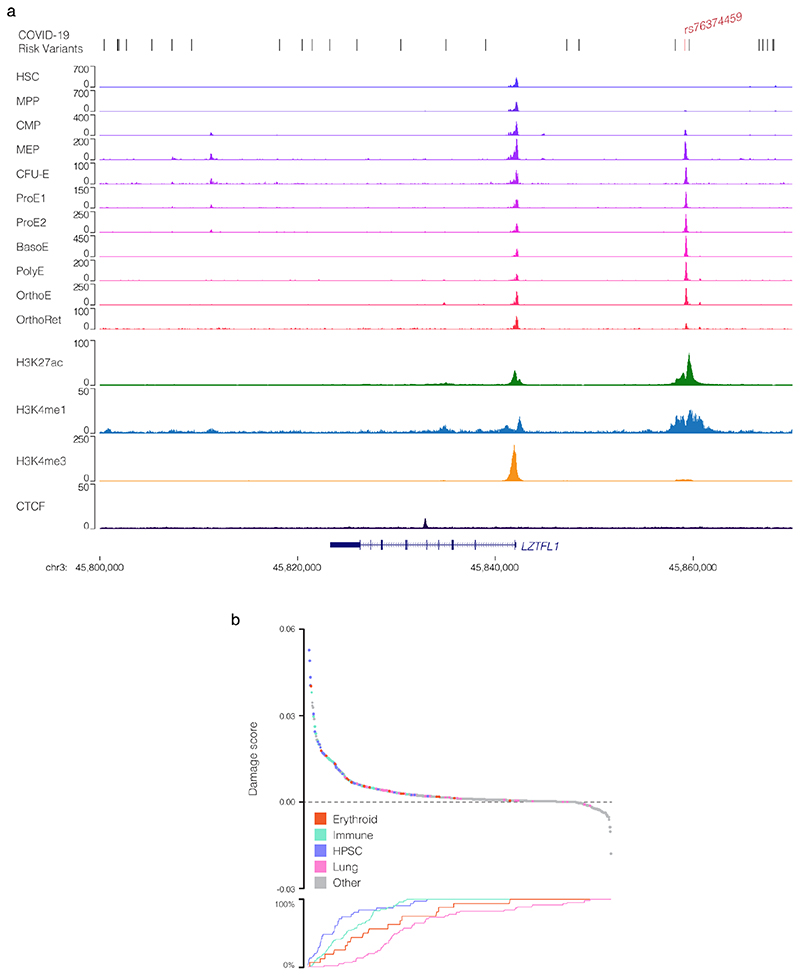
Extended Data Figure 4. rs76374459 is likely benign in an erythroid enhancer.
ATAC-seq from progenitor80 and differentiating erythroid cells81. Haematopoietic Stem Cells (HSC), Multi-Potent Progenitors (MPP), Common Myeloid Progenitors (CMP), Myeloid-Erythroid Progenitors (MEP) from bone marrow or peripheral blood and erythroid Colony Forming Units (CFU-E), Pro-erythroblasts (ProE1, ProE2), Basophilic Erythroblasts (BasoE), Polychromatic Erythroblasts (PolyE), Orthochromatic Erythroblasts (OrthoE) and Orthochromatic/Reticulocytes (OrthoRet). ChIP-seq tracks from CD71+ CD23+ mature erythroid cells16 show presence of marks associated with active transcription (H3K27ac), enhancers (H3K4me1), promoters (H3K4me3) and boundaries (CTCF). b, deepHaem damage score for the risk-C allele versus non-risk-G allele of rs76374459 associated with severe COVID-19 in 694 cell-types. rs763774458 is found in open chromatin through-out erythropoiesis. A positive score predicts loss of accessibility, a negative score predicts increased accessibility.