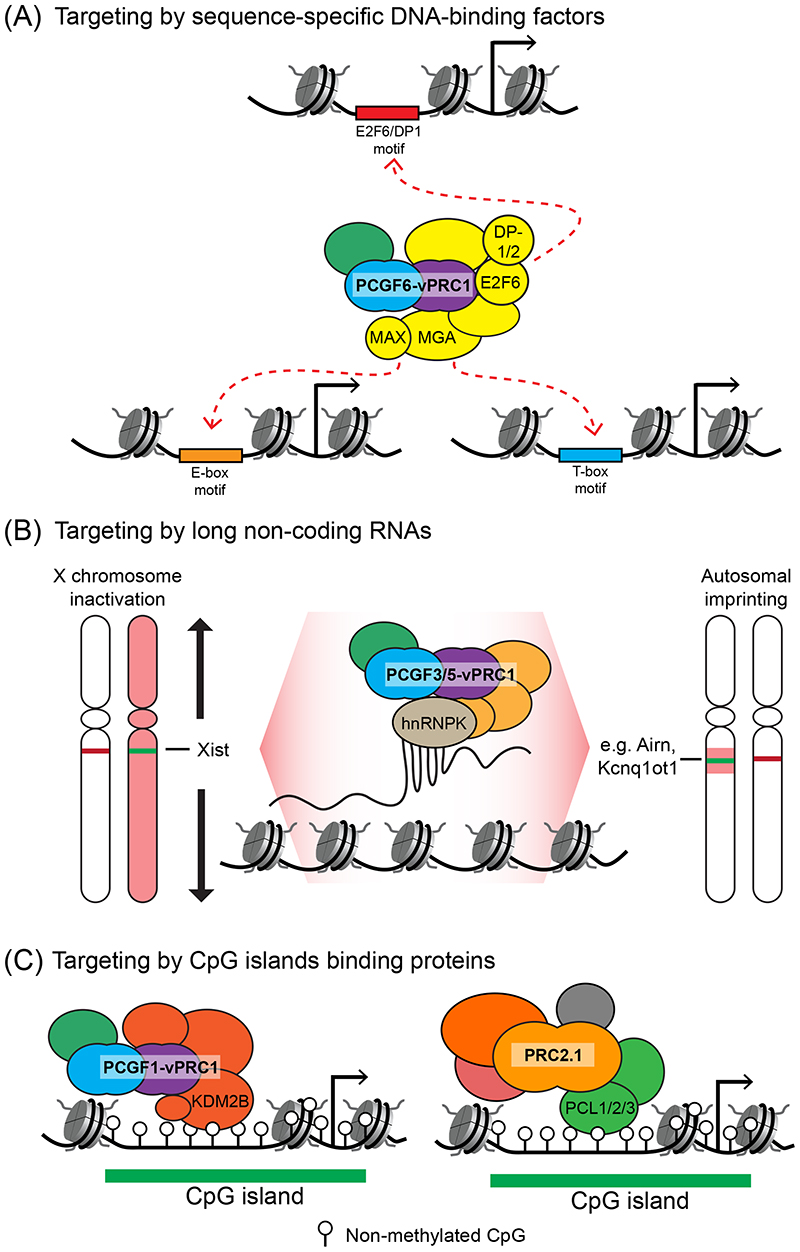
Figure 2. Primary mechanisms of Polycomb target-site identification.
(A) Sequence-specific DNA-binding factors in the Polycomb group RING finger 6 (PCGF6)-containing variant Polycomb repressive complex 1 (vPRC1) complex recognise DNA sequence motifs at target sites. These DNA-binding factors include MAX gene-associated (MGA)–MYC-associated factor X (MAX) and E2F6–dimerization partner 1 or 2 (DP-1/2) dimers, which bind to different DNA sequence motifs and contribute to sequence-specific PCGF6-vPRC1 targeting in different contexts.
(B) Polycomb complexes can identify chromosomal binding sites during X chromosome inactivation through the long-noncoding RNA (lncRNA) X inactivation specific transcript (XIST); in autosomal imprinted regions mono-allelic gene repression is achieved through the lncRNAs Airn and Kcnq1ot1. In these regulatory contexts, the adaptor protein heterogeneous nuclear ribonucleoprotein K (hnRNPK) is thought to interact with the lncRNA and recruit the PCGF3/5-vPRC1 complex. These mono-allelic targeting cases are atypical in that they nucleate the binding of Polycomb complexes at large chromosomal regions, whereas Polycomb complex targeting to the majority of genomic sites is more punctate and associated with gene regulatory elements (see parts A and C).
(C) Both PCGF1-vPRC1 and PRC2.1 are targeted to CpG islands. The lysine-specific demethylase 2B (KDM2B) subunit of PCGF1-vPRC1 contains a zinc finger-CxxC (ZF-CxxC) domain that binds specifically to non-methylated CpG dinucleotides. In PRC2.1, the Polycomb-like 1 (PCL1), PCL2 or PCL3 subunit contains a winged-helix domain that binds non-methylated CpG dinucleotides in certain sequence-contexts.