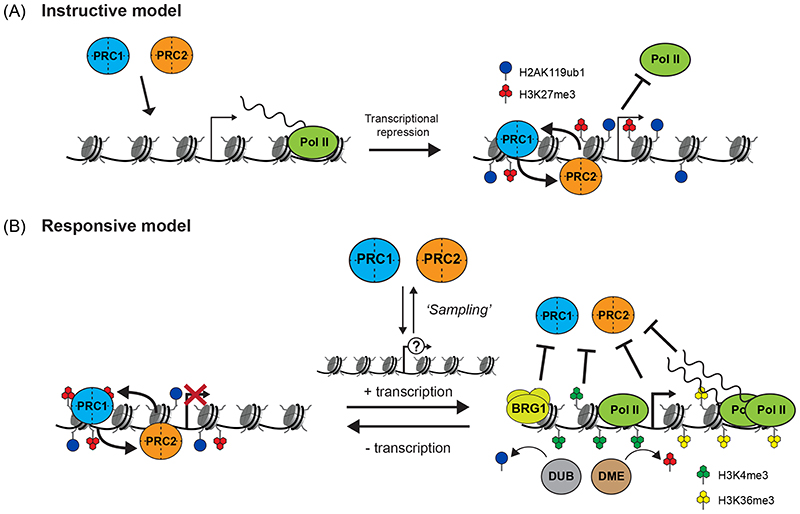
Figure 5. Models of Polycomb chromatin domain formation and gene regulation.
(A) An instructive model, in which Polycomb complexes are recruited to target sites (left), where they initiate the formation of Polycomb chromatin domains (right) and drive the repression of transcribed genes.
(B) A responsive model, in which Polycomb complexes dynamically ‘sample’ potential target sites for susceptibility to Polycomb chromatin domain formation. In particular, the Polycomb group RING finger 1 (PCGF1)-containing variant Polycomb repressive complex 1 (vPRC1) and PRC2.2 complex, through their capacity to bind CpG islands (not shown), could dynamically engage with approximately 70% of gene promoters. At lowly or untranscribed genes (left), these complexes could potentially sense and respond to the (near) absence of transcription by ubiquitylating histone H2A Lys119 (H2AK119ub1) and tri-methylating histone H3 Lys27 (H3K27me3) to initiate the formation and spreading of Polycomb chromatin domains, which could help counteract low-level or inappropriate transcription and maintain an inactive chromatin state to protect cell identity. However, at expressed genes (right), transcription-associated features including H3K4me3 and H3K36me3, high levels of nascent transcripts, BRG1-mediated chromatin remodelling, and deubiquitylase (DUB) and demethylase (DME) activities, counteract H2AK119 and H3K27 modification, thereby blocking Polycomb chromatin domain formation and limiting Polycomb function at these genes.
Pol II, RNA polymerase II.