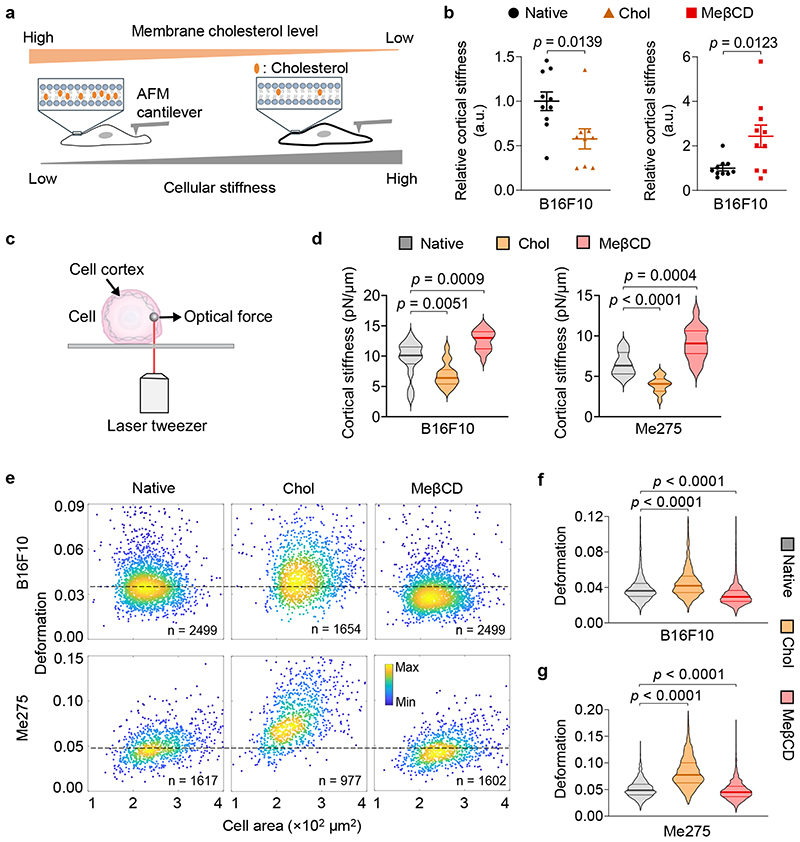
Fig. 2. Cancer-cell stiffness can be manipulated via the supplementation or depletion of cholesterol in the cell membrane.
a, Schematic illustration of the correlation between cellular stiffness and membrane cholesterol level. b, Relative cortical stiffness determined by nanoindentation measurements using atomic force microscopy (AFM) for native, Chol- or MeβCD-treated B16F10 cancer cells (n = 9 ~ 10 individual cells). Each data point is the average of at least twenty force curve measurements of a single cancer cell. Native B16F10 cancer cells serve as a standard (100%). Error bars represent SEM. c, Schematic illustration of the optical tweezer setting for cell cortical stiffness measurement. d, Cortical stiffness of native, Chol- or MeβCD-treated murine B16F10 and human Me275 cancer cells measured by the optical tweezer (n = 14 ~ 17 individual cells). e-g, Cellular deformation was measured using deformability cytometry to compare cellular stiffness in a high throughput manner. Shown are representative scatter plots (e; indicated are sample size, outliers not shown) and quantitative deformation of native, Chol- or MeβCD-treated murine B16F10 (f) and human Me275 (g) cancer cells. In all the violin plots (d, f, g), the middle solid line shows median, and lower and upper dash lines show 25th and 75th percentiles, respectively. P values were determined by unpaired Student’s t test. a.u., arbitrary unit; n.s., not significant.