Abstract
The ability to genetically engineer pathogenic mycobacteria has increased significantly over the last decades due to the generation of new molecular tools. Recently, the application of the Streptococcus pyogenes and the Streptococcus thermophilus CRISPR-Cas9 systems in mycobacteria has enabled gene editing and efficient CRISPR interference-mediated transcriptional regulation. Here, we converted CRISPR interference into an efficient genome editing tool for mycobacteria. We demonstrate that the Streptococcus thermophilus CRISPR1-Cas9 (Sth1Cas9) is functional in Mycobacterium marinum and Mycobacterium tuberculosis, enabling highly efficient and precise DNA breaks and indel formation, without any off-target effects. In addition, with dual sgRNAs this system can be used to generate two indels simultaneously or to create specific deletions. The ability to use the power of the CRISPR-Cas9-mediated gene editing toolbox in M. tuberculosis with a single step will accelerate research into this deadly pathogen.
Keywords: CRISPR-Cas9 system, genome editing, indels, Mycobacterium marinum, Mycobacterium tuberculosis
Abbreviations
- CRISPR
clustered regular interspaced short palindromic repeat
- CRISPRi
CRISPR interference
- Sth1Cas9
Streptococcus thermophilus CRISPR1-CRISPR-associated protein 9
- sgRNA
single guide RNA
- NHEJ
non-homologous end joining
- HR
homologous recombination
- ATc
anhydrotetracycline
- indel
insertion or deletion
- PAM
protospacer adjacent motif
- INH
isoniazid
1. Introduction
Mycobacterium tuberculosis remains the world’s leading infectious agent, killing 1.5 million people annually [1]. In order to increase our understanding of the pathogenesis of tuberculosis and to identify and optimize treatment and prevention methods, efficient genetic tools to manipulate the mycobacterial genome are highly desired.
The clustered regular interspaced short palindromic repeat (CRISPR)-technology has revolutionized the ability to edit genomes. Since the discovery of the class II CRISPR systems, the number of species that have been genetically modified by this technique continues to expand. The CRISPR-associated protein 9 (Cas9), containing two endonuclease domains, forms a complex with a single guide RNA (sgRNA) designed to recognize a target sequence of interest, located next to a protospacer adjacent motif (PAM) sequence. Upon recognition, a precise double-stranded DNA break is induced [2], which is detrimental for survival if not repaired. Distinct DNA repair mechanisms of the host cell, including non-homologous end joining (NHEJ) or homologous recombination (HR) repair mechanisms, will result in repair of the cleaved DNA strands. The NHEJ repair mechanism is error-prone and is likely to result in random insertions and deletions (indels) and subsequent frameshift mutations [3, 4]. Most bacteria lack a NHEJ repair system and therefore cannot efficiently repair double-stranded DNA breaks resulting in bacterial cell death when the CRISPR-Cas9 system is applied. However, mycobacteria do contain such a DNA repair system [5–7], which was previously used to produce indel mutations and gene deletions in Escherichia coli by heterologous expression of the mycobacterial NHEJ repair proteins Ku and Ligase D (LigD) [8]. Thus far, CRISPR-assisted mycobacterial genome editing was achieved in Mycobacterium smegmatis utilizing a codon-optimized Cfp1 from Corynebacterium glutamicum [9]. Additionally, CRISPR-Cas-assisted NHEJ gene editing was recently accomplished in M. smegmatis, M. marinum and M. tuberculosis by inhibition of RecA-dependent HR-mediated repair in addition to overexpression of mycobacterial NHEJ proteins [10].
Besides gene editing, the CRISPR-Cas9 system has been adapted to several other approaches, including CRISPR interference (CRISPRi) to inhibit gene expression, by the creation of a catalytically defective version of the Cas9 protein (dCas9), sometimes fused to a transcriptional inhibitor [11, 12]. CRISPRi technology has been successfully applied to many organisms, including mycobacterial species [13, 14]. Although CRISPRi is a powerful tool, especially for essential genes, it requires optimization and validation for every targeted gene and has strong polar effects on genes in the same operon. Moreover, repression of protein production is not always suitable, since a small fraction of remaining protein can be sufficient for its full biological function [13]. In this work, we expanded the genetic toolbox for mycobacteria by restoring the enzymatic activity of catalytically inactive Streptococcus thermophilus CRISPR1-Cas9 (Sth1dCas9) that is widely used for CRISPRi. We demonstrate that, by using a single plasmid containing active Sth1Cas9, precise and efficient gene editing can be achieved in mycobacterial species without the need to introduce NHEJ proteins or to inhibit RecA-dependent HR-mediated repair. The power of this one-step gene-editing tool in mycobacteria will accelerate research into this important group of pathogens.
2. Materials and reagents
2.1. Strains, media and cell growth conditions
All strains used in this study were derived from the wild-type strains Mycobacterium marinum M [15] or E11 [16] and Mycobacterium tuberculosis strains CDC1551 [17], H37Rv [18] and mc26020 (ΔLysA ΔpanCD) [19, 20]. All strains were routinely cultured on Middlebrook 7H10 plates supplemented with OADC (Difco) or in Middlebrook 7H9 medium (Difco) containing ADC (Difco) and 0.05% Tween-80 (Sigma). For the culture of M. tuberculosis mc 2 6020 (ΔLysA ΔpanCD), 100 ng/ml L-lysine (Sigma) and 25 ng/ml D-panthotenic acid (Sigma) were supplemented. When required, the appropriate antibiotic was added (50 μg/ml kanamycin (Sigma), 50 μg/ml hygromycin (Roche) or 30 μg/ml streptomycin (Sigma). M. marinum and M. tuberculosis cultures and plates were incubated at 30°C and 37°C, respectively. E. coli TOP10F' and DH5α were used for cloning experiments to generate the plasmids pCRISPRx-Sth1Cas9-L5 and pTdTomato-L5, and were grown at 37°C on LB agar plates and in LB medium. When necessary, antibiotics were added to the cultures or plates at similar concentrations as for M. marinum cultures. Competent mycobacteria were made as follows: M. marinum and M. tuberculosis were grown in 7H9 medium supplemented with ADC and 0.05% Tween-80. At OD600 0.8-1.5, 1.5% glycine was added to the culture and incubated for 16 hours. Cells were harvested and washed three times with (cold) 10% glycerol prior to resuspending the bacteria at a concentration of 50 OD/ml in 10% glycerol.
2.2. Target site prediction
The genomes of M. marinum (ASM1834v1) and M. tuberculosis (ASM19595v2) were converted to GFF3 and added to the CHOPCHOP web interface. Targets were predicted using CHOPCHOP [21]. The custom PAM sequences ‘NNAGAA’, ‘NNGGAA’ and ‘NNAGCAT’ were used to identify target sites. When the 5’ of the target site was not equal to guanine, the target site was extended to the first guanine present in the genome. Potential sgRNAs containing protospacers with less than 4 mismatches at other loci in the genome were labeled as potential off-targets and excluded.
2.3. Plasmids
PLJR962 [13] was used as a template to perform extension PCR to reintroduce the catalytic residues in Sth1Cas9 (see primer table for sequences). The PCR product was cloned into pJet1.2 (ThermoFisher) and sequence verified by Sanger sequencing (Macrogen). Sth1Cas9 was cut from pJET1.2 and cloned into PLJR962, both digested with restriction enzymes SbfI-ClaI, generating pCRISPRx-Sth1Cas9-L5 (Addgene 140993).
All oligo’s encoding sgRNAs used in this study are listed in Supplementary Table 1. Oligo’s contained BsmBI overhangs as described in [13]. Annealed oligos were ligated into the pCRISPRx-Sth1Cas9-L5 plasmid at BsmBI restriction sites, as described previously [13].
For the incorporation of sgRNA-crtb-1 and sgRNA-crtb-2 in the same plasmid, pCRISPRx-Sth1Cas9-L5 plasmid was digested with NdeI and BamHI. Restriction overhangs were blunted by Klenow fragment (ThermoFisher) prior to ligation to create an intermediary plasmid. sgRNA-crtb-2 was incorporated into the intermediate plasmid through BsmBI restriction sites, amplified by PCR and ligated into pCRISPRx-Sth1Cas9-L5 containing sgRNA-crtb-1 through BspQI restriction and insertion cloning.
For the generation of pTdTomato-L5, the gene encoding TdTomato was amplified using PCR from pML2424 [22] utilizing the primers described in Supplementary Table 2. The PCR product was cloned using In-Fusion Cloning (Takara, 639650) into pMV361-eccB5-mycP5-HA (strepR) [23] digested with EcoRI and HindIII, generating the plasmid pTdTomato-L5 (Addgene 140994).
2.4. Generation and identification of mutants
One hundred microliter of electrocompetent bacteria were electroporated with 1 μg of pCRISPRx-Sth1Cas9-L5 plasmid containing the appropriate single or double sgRNA(s) with a single pulse of 2.5 kV, 25 μF, 720 Ω. Subsequently, bacteria were either plated on 7H10 plates containing kanamycin, with or without 100 ng/ml ATc (IBA Lifesciences), or bacteria were shortly induced with 100 ng/ml ATc for 1h after 5h recovery in 7H9 medium and plated on 7H10 plates without ATc. Following incubation for 7-10 days at 30°C, separate colonies were picked for PCR amplification using Phusion High Fidelity DNA polymerase (ThermoFisher) and sequencing analysis (Macrogen). Primers used in this study are listed in Supplementary Table 2. The Synthego ICE analysis tool was utilized to identify the presence of indels in mixed sequencing samples. pCRISPRx-Sth1Cas9-L5 plasmids containing sgRNA targeting PE_PGRS35 (Rv1983, MT2036) or PE_PGRS16 (Rv0977, MT1004) were electroporated into M. tuberculosis H37Rv and CDC1551 strains. pCRISPRx-Sth1Cas9-L5 plasmid containing sgRNA targeting katG (Rv1908c) was electroporated into M. tuberculosis H37Rv mc26020 strain, with a single pulse of 2.5 kV, 25 μF, 1000 Ω. Following overnight recovery in fresh medium, bacteria were plated on 7H10 plates containing OADC and 50 μg/ml kanamycin and incubated at 37°C for three weeks. Subsequently, bacteria were picked and grown on fresh 7H10 plates containing 100 ng/ml ATc. Following incubation of three weeks, separate colonies were picked and DNA was isolated for PCR and sequencing analysis.
2.5. Replacement of CRISPR-Cas9 machinery
pTdTomato-L5 was electroporated into a crtB mutant strain obtained with sgRNA-crtb-1 and a crtB gene deletion mutant acquired using pCRISPRx-Sth1Cas9-L5 containing sgRNA-crtb-1 and sgRNA-crtb-2 and grown for 7-10 days at 30°C on 7H10 plates containing 30 μg/ml streptomycin. Separate colonies were diluted in 100 μl phosphate-buffered saline (PBS). Subsequently, 10 μl was spotted on plates containing either streptomycin or kanamycin.
2.6. Assessment of INH resistance
M. tuberculosis H37Rv mc26020 strains carrying the pCRISPRx-Sth1Cas9-L5 plasmid with sgRNA targeting katG were diluted in PBS and 10 μl was spotted on 7H10 plates containing isoniazid (INH; Sigma) in a concentration of 0 μg/ml, 0.019 μg/ml, 0.056 μg/ml and 0.19 μg/ml and incubated at 37°C for 3 weeks. Colony density was measured using ImageJ dot plot analysis. For each strain, the integrated density of different INH concentrations was normalized to the 0 μg/ml INH control.
2.7. Whole-genome sequencing
DNA was isolated from M. marinum M WT (parent strain) and crtB mutants, obtained using sgRNA-crtb-1 following short (1h) and long ATc treatment (10 days). Bacteria were harvested from 7H10 plates and disrupted in a 1:1 mixture of TE buffer and phenol/chloroform/iso-amylalcohol (50:49:1) using 0.1 mm Zirconia silicon beads (Biospec Products) in a bead beater for 1 min. Following centrifugation, the aqueous phase was incubated with isopropanol and 300 mM sodium acetate for 15 minutes at room temperature. After centrifugation, the pellet was washed with 70% ethanol and dried prior to reconstitution in TE buffer and RNAse A (ThermoFisher) treatment. DNA was further isolated using the High pure PCR preparation kit (Roche). Whole-genome sequencing and further preparation was performed by Novogene (United Kingdom). Briefly, DNA was sheared into fragments of 350 bp, and library was constructed utilizing the NEBNExt DNA Library Prep Kit. Concentration and insert size were determined by Qubit 2.0 fluorometer and Agilent 2100 bioanalyzer prior to Illumina high-throughput sequencing. Coverage and indel analysis were performed utilizing QIAGEN CLC Genomics Workbench. Indels were considered true when the number of reads was more than 10.
2.8. Calculations
Percent survival was calculated as the number of colonies on plates transformed with pCRISPRx-Sth1Cas9-L5 plasmids containing a targeting sgRNA compared to the number of colonies containing a non-targeting sgRNA. Percentages of phenotypic crtB mutants were calculated as the percentage of white colonies compared to the total number of colonies. Gene editing efficiency is defined as the percentage of mutated colonies compared to the total number of colonies successfully sequenced. Frameshift editing efficiency is defined as the percentage of colonies containing a frameshift mutation compared to the total number of colonies successfully sequenced. Percentages are first calculated on a per gene basis, followed by calculating the average of these percentages when multiple genes are involved. Only sequence-verified samples are included. Colonies for which no Sanger sequence could be obtained are excluded.
3. Results
3.1. CRISPR-Cas9-assisted gene editing in M. marinum
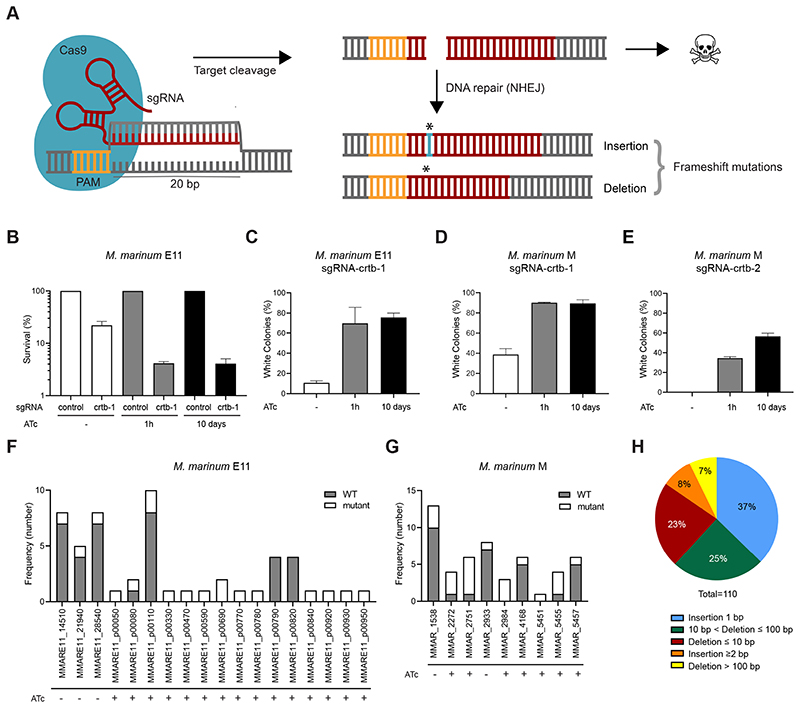
In order to achieve gene editing in mycobacteria, we reverted the previously described catalytically inactive Sth1dCas9 used in CRISPRi [13] into a functional endonuclease. Sth1dCas9 was modified using site-directed mutagenesis by mutating alanine 9 into aspartic acid (A9D) and alanine 599 into a histidine (A599H) on the L5-integrative plasmid PLJR962 to create a functional Sth1Cas9, expressed under the control of an anhydrotetracycline (ATc)-inducible promoter. We designated this plasmid pCRISPRx-Sth1Cas9-L5. Restoring the endonuclease function of Sth1Cas9 enables the generation of double-stranded breaks in the bacterial chromosome in the presence of specific sgRNA (Figure 1A).
Figure 1. Gene editing using CRISPR-Cas9 in M. marinum.
(A) Schematic overview of CRISPR-Cas9-mediated gene editing. Following induction with ATc, Sth1Cas9 and the designed sgRNA are expressed and form a complex. Following the recognition of the target site next to the PAM, the endonuclease domains of Sth1Cas9 will induce a double-stranded DNA break which can result in bacterial killing if not repaired. NHEJ results in error-prone DNA repair, which can lead to frameshift mutations and subsequent knockout of the target gene. (B) Survival of M. marinum E11 transformants carrying pCRISPRx-Sth1Cas9-L5 containing sgRNA-crtb-1. Survival was calculated as the percentage of the total number of colonies compared to the number of colonies carrying the control plasmid. Bars represent the mean values and standard deviations from two experiments. (C-E) Percentages of phenotypically white colonies using sgRNA-crtb-1 in M. marinum E11 (C) and M (D) strains, and sgRNA-crtb-2 in M. marinum M (E). Percentages of phenotypically white colonies were calculated as the percentage of white colonies in proportion to the total number of colonies. Mean and standard deviation are shown from two replicate experiments and three replicate experiments for sgRNA-crtb-1 in M. marinum M. (F) Frequency of indel mutants for several genes in M. marinum E11. Separate colonies with or without ATc treatment were screened for indels using target amplification and sequencing analysis. The number of colonies harboring indels at the target site are shown in white, and WT colonies are shown in gray. (G) Number of colonies containing CRISPR-Sth1Cas9-induced mutations for several genes tested in M. marinum M as identified by sequencing. Separate colonies were subjected to genome target amplification and sequencing analysis. WT colonies are shown in gray, and mutant colonies are shown in white. (H) Frequency of mutation events observed for all mutated colonies in M. marinum E11 and M strains as determined by PCR and subsequent sequencing analysis for the indicated genes.
To explore the gene-editing capacity of this CRISPR-Cas9-mediated system in M. marinum, sgRNAs were designed to target the non-essential crtB gene. CrtB is required for the production of a yellow carotenoid pigment in the presence of light. Therefore, non-functional crtB mutants are easily distinguished as white colonies in contrast to the yellow wild-type (WT) M. marinum colonies [24]. The pCRISPRx-Sth1Cas9-L5 plasmid containing a sgRNA that targets crtB (sgRNA-crtb-1) was transformed into WT M. marinum M and E11 strains. As expected, we obtained a reduced number of transformants (23%) with our targeting construct in comparison to transformants carrying the control vector containing a non-targeting sgRNA (Figure 1B; Supplementary Table 3). This difference is likely due to bacterial cell death after induction of double-stranded DNA breaks. Moreover, the number of transformants carrying a functional CRISPR-Cas9 system decreased to 4% following induction of the CRISPR-Cas9 system, either when the inducing agent ATc was added to the growth medium of a plate (for 10 days) or when it was only briefly added (for 1h) during recovery in liquid medium following electroporation. Next, we investigated whether genome editing occurred in the transformants carrying the functional CRISPR-Cas9 system. Following electroporation of the pCRISPRx-Sth1Cas9-L5 harboring sgRNA-crtb-1, 12% and 42% of the colonies showed the phenotype of crtB mutants in M. marinum E11 and M strains, respectively (Figure 1C, D, Supplementary Table 3). This demonstrates that CRISPR-Cas9-assisted gene editing was already successful in the absence of ATc induction, probably due to leaky expression of the CRISPR-Cas9 system. Upon ATc induction, the levels of non-functional crtB mutants were significantly increased to 66-90% (Figure 1C, D, Supplementary Figure 1A). This effect was observed both following ATc treatment for 1h during electroporation recovery in liquid medium or for 10 days on plates. These experiments indicate that ATc-induced CRISPR-Cas9-mediated gene editing is highly efficient.
Previous work identified that, next to the consensus PAM sequence NNAGAAW for Sth1Cas9 [25, 26], some divergence of this PAM sequence is tolerated depending on Sth1Cas9 concentration [27]. Since mycobacterial DNA is GC-rich, the number of consensus PAM options in the genome are limited. Therefore, we examined whether NNGGAA could also be used as a PAM sequence to direct gene editing. Again, we targeted the crtB gene as a marker gene for inactivation using sgRNA-crtb-2. Transformants carrying the pCRISPRx-Sth1Cas9-L5 plasmid containing sgRNA-crtb-2 in the absence of ATc induction did not yield any non-functional crtB mutants (Figure 1E), indicating that gene editing using sgRNA-crtb-2 might require induction of the CRISPR-Cas9 system. Indeed, upon treatment with ATc, either for 1h or 10 days, 35% and 55% of the colonies showed the crtB mutant phenotype in M. marinum M, respectively. This demonstrates that a suboptimal PAM site is suitable for creating mutants, although with a lower efficiency as compared to targeting the optimal PAM sequence. Gene editing with the sgRNA-crtb-2 might not be feasible for the M. marinum E11 strain, as it harbors 3 mismatches within the 20 bp sgRNA target site. We investigated whether this mismatch would abolish the generation of non-functional crtB mutants. Indeed, no white colonies were detected in M. marinum E11 strain following ATc induction, demonstrating that the editing system does not tolerate these 3 mismatches (data not shown). Thus far, we used visual selection to identify mutants. To establish what type of mutations were generated in these non-functional crtB mutants, we also performed PCR amplification combined with Sanger sequencing. Analysis of the amplified genomic target regions revealed that small random insertions and deletions (indels), both in-frame (27%) and out-of-frame (73%), occurred at the location of the sgRNA target site. Apparently, in-frame mutation events in the targeted regions also result in loss of function for CrtB. Since disruption of the crtB gene works very efficiently, we also used our procedure to disrupt other genes in M. marinum. We targeted genes coding for ESAT-6 secretion system-1 (ESX-1)-specific proteins EspB (MMAR_5457), EspD (MMAR_4168), EspI (MMAR_5451), EspK (MMAR_5455), a possible TetR family transcriptional regulator (MMAR_2984), Lon protease (MMAR_2751), and (putative) aspartic proteases PecA (MMAR_2933, MMARE11_28540; [28]) PE_PGRS16 (MMAR_2272, MMARE11_21940) and PE26 (MMAR_1538, MMARE11_14510; Figure 1F, G). In addition, we targeted genes located on the large natural plasmid pRAW [29] in M. marinum E11 (Figure 1F). In total, 27 different genes were targeted using CRISPR-Cas9 technology, often combined with ATc induction. We examined a small number of colonies for all targets by amplifying the region of interest followed by Sanger sequencing. Thus far, using this procedure, CRISPR-Cas9-induced indel events were identified for 25 out of 27 genes targeted (Supplementary Table 4), with a gene editing efficiency of 63.6%. Frameshift indels, which are desired for loss of function, were observed in 23 out of 27 genes with an average frameshift editing efficiency of 55%. In-frame mutations accounted for 10% of the mutant colonies tested. Most CRISPR-Cas9-induced mutations observed were 1 bp insertions, followed by (small) deletions up to 100 bp. Moreover, occasionally large deletions of >100 bp or insertions ≥2 bp were detected. (Figure 1H). The majority of the mutants were generated using the consensus PAM sequence NNAGAAW. Furthermore, frameshift mutants were identified following targeting with PAM sequences NNGGAA as well as NNAGCAT, further highlighting that divergence from the consensus Sth1Cas9 PAM sequence is possible for CRISPR-Cas9-mediated gene editing in M. marinum (Supplementary Table 4). This analysis shows that the production of indels in M. marinum with Sth1Cas9 system is efficient.
3.2. Gene editing using the CRISPR-Cas9 system does not induce off-target mutations
The fragment encoding the CRISPR-Cas9 machinery utilized in our procedure is integrated into the attB site on the mycobacterial genome. Although the Sth1Cas9 endonuclease is under control of an ATc-inducible promoter, we observed that mutants were already generated without ATc induction. Therefore, the continuous, low-level expression of Sth1Cas9 could theoretically induce additional DNA mutations after prolonged culturing. To overcome this potential problem, another L5-integrative plasmid was constructed to replace the plasmid carrying the CRISPR-Cas9 system. This integrative pMV361-based construct, encoding a streptomycin resistance gene and the fluorescent protein tdTomato under the constitutive promoter hsp60 (pTdTomato-L5), was electroporated into crtB mutants. All colonies tested were tdTomato-positive and streptomycin-resistant. None of these colonies retained kanamycin resistance, indicating that the CRISPR-Cas9 cassette was efficiently replaced (Supplementary Figure 1B).
Although replacing the CRISPR-Cas9 system with another integrative vector reduces the chance of off-target effects, some off-target mutations could already have been induced. To test this, we performed whole-genome sequencing on two crtB mutants obtained using sgRNA-crtb-1 following either 1h or 10 day ATc treatment and their parent strain. Average read depths of 224x, 302x and 283x were obtained for the WT parent strain, 1h and 10 day ATc conditions, respectively. We analyzed the prevalence of additional indels in the genomes of the crtB mutants in comparison with the parental strain. We found that, apart from the deletions at the crtB target site, not a single additional indel was introduced in the presence of the CRISPR-Cas9 system, which shows that, with this knockout construct, even following prolonged induction (10 days), additional genomic mutations are not common.
3.3. CRISPR-Cas9-assisted gene editing in M. tuberculosis
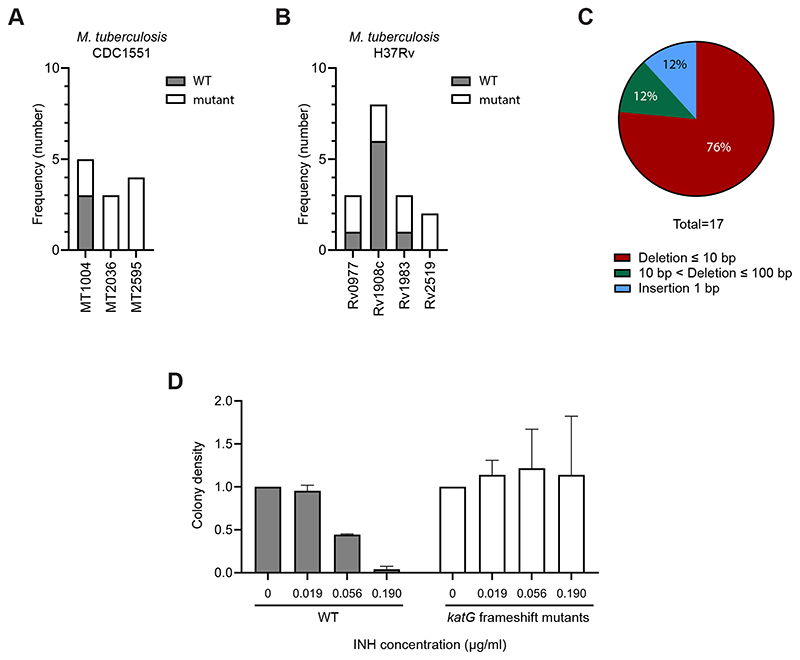
Since CRISPR-Cas9-mediated gene editing is highly efficient in M. marinum, we investigated whether the same system could be employed in M. tuberculosis. The pCRISPRx-Sth1Cas9-L5 plasmids containing sgRNA targeting the genes coding for aspartic proteases PecA, also known as PE_PGRS35 (Rv1983, MT2036)[28], PE_PGRS16 (Rv0977, MT1004) and PE26 (Rv2519, MT2595) were transformed into M. tuberculosis strains H37Rv and CDC1551. Following ATc induction on plates, gene fragments from separate colonies were amplified by PCR. Sequencing analysis revealed the identification of frameshift mutants for all 7 genes targeted, with a gene editing efficiency of 71% and a frameshift editing efficiency of 52% (Figure 2A-B; Supplementary Table 4). Additionally, 23.5% of the mutant colonies tested showed an in-frame mutation event. In M. tuberculosis, predominantly small deletion events up to 12 bp occurred at the cleavage site, in addition to 1 bp insertions (Figure 2C). Moreover, consistent with gene editing in M. marinum, mutation events in M. tuberculosis were also achieved using the less efficient PAM sequences NNGGAA and NNAGCAT, which indicates the flexibility of possible target sites in the genome of M. tuberculosis (Supplementary Table 4).
Figure 2. CRISPR-Cas9 mediated gene editing in M. tuberculosis.
(A) The number of colonies harboring indel mutations in M. tuberculosis CDC1551. Transformants carrying the pCRISPRx-Sth1Cas9-L5 containing sgRNA targeting either PE_PGRS16 (MT1004), pecA (MT2036) or PE26 (MT2595) were induced on 7H10 plates containing ATc for 3 weeks. Separate colonies were tested for mutations at the sgRNA target sites using PCR and sequencing. WT colonies are shown in gray, and mutant colonies are shown in white. (B) The number of colonies containing indel mutations in M. tuberculosis H37Rv. The pCRISPRx-Sth1Cas9-L5 containing sgRNA targeting either katG (Rv1908c), PE_PGRS16 (Rv0977), pecA (Rv1983) or PE26 (Rv2519) were electroporated in M. tuberculosis H37Rv. Genome editing was induced for 3 weeks on 7H10 plates supplemented with ATc. Mutants were identified using target amplification and sequencing. Colonies identified as WT are shown in gray, and mutant colonies are shown in white. (C) Frequency and type of indels observed at sgRNA target sites among the mutants detected. (D) Isoniazid (INH)-resistance of katG frameshift mutants. Two WT and katG frameshift mutant strains were spotted on 7H10 plates containing different concentrations of INH. Colony density was measured using ImageJ analysis. Data for each strain was normalized to the untreated condition.
To assess the functionality of CRISPR-Cas9-generated frameshift mutants in M. tuberculosis, we targeted the gene coding for KatG, the catalase-peroxidase enzyme that is responsible for activating the prodrug isoniazid (INH) in M. tuberculosis [30]. Following induction of the CRISPR-Cas9 machinery, two katG frameshift mutants were identified by target amplification and Sanger sequencing. Since the prodrug is not activated in the absence of KatG, we analyzed whether our frameshift mutants gained resistance to INH. Therefore, WT and katG frameshift mutants were incubated on plates containing different concentrations of INH. Whereas bacterial growth was severely inhibited for the WT strains, colony densities of the katG frameshift mutants were not affected (Figure 2D; Supplementary Figure 1C). These results demonstrate that CRISPR-Cas9-induced frameshift mutations abolish protein function of the target gene.
3.4. CRISPR-Cas9-mediated gene deletions in M. marinum
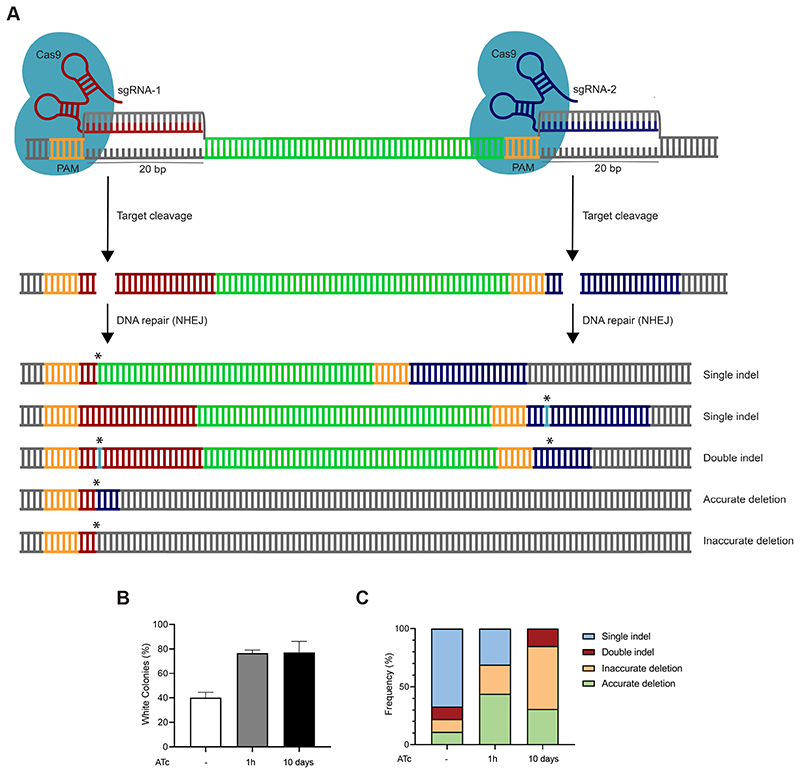
In our assays, the CRISPR-Cas9-mediated system enabled the efficient generation of frameshift mutants in M. marinum and M. tuberculosis. Next, we sought to explore whether specific deletions could be achieved by the simultaneous introduction of two sgRNAs targeting the same gene and precisely deleting the DNA fragment between them (Figure 3A). Therefore, the pCRISPRx-Sth1Cas9-L5 plasmid containing both sgRNA-crtb-1 and sgRNA-crtb-2 was introduced into M. marinum M. The target DNA positions of these dual sgRNAs were separated by 379 bp within the crtB gene. In line with our previous results using sgRNA-crtb-1, following the introduction of the dual sgRNA pCRISPRx-Sth1Cas9-L5 plasmid, 41% of the transformants already appeared non-functional crtB mutants without the addition of ATc (Figure 3B; Supplementary Table 3). The amount of phenotypic crtB mutants was further enhanced to 76% and 72% following 1h or 10 day ATc treatment, respectively. Next, white colonies were analyzed for the presence and prevalence of gene fragment deletions. In the absence of ATc, the CRISPR-Cas9 system mainly yielded single indels, where mutations were positioned at either one of the sgRNA target sites (67%). Moreover, cases of double indels, with mutations at both sgRNA target sites (11%) or gene fragment deletions were observed, removing the gene fragment between the two sgRNA sites (22%) (Figure 3C; Supplementary Table 5). In all cases, these gene fragment deletions were accompanied by indels at the predicted cleavage sites. DNA deletions that were located between the 20 bp sgRNA target sites were considered as accurate deletions, while the deletions extending beyond the 20 bp sgRNA sites were regarded as inaccurate deletions. Without ATc induction, we observed that 11% of the colonies contained accurate deletions, and 11% harbored inaccurate deletions. Upon induction of the CRISPR-Cas9 machinery for 1h or 10 days, a decrease in the prevalence of single indels was observed (31% and 0%). Interestingly, although double indels were not detected following short ATc induction, they were observed in 15% of the colonies tested following ATc induction on plates. More importantly, gene fragment deletions were also increased to 69% and 85% following short and prolonged ATc treatment, respectively. Accurate deletions were detected for 44% of the phenotypic crtB mutants obtained with 1h ATc induction and 31% of the mutants obtained with 10 day ATc induction on plates (Figure 3C). However, an increased amount of inaccurate deletions were observed for 25% and 54% after brief and long ATc treatment, respectively. These experiments indicate that specific deletions can be efficiently generated within a gene or a particular region of the mycobacterial genome by simultaneously expressing two sgRNAs. However, these deletions are not always accurate and can sometimes extend beyond the target sites.
Figure 3. CRISPR-Cas9-mediated gene deletions using dual sgRNAs in M. marinum.
(A) Schematic overview of CRISPR-Cas9-induced gene deletions using dual sgRNAs. Mycobacteria carrying the pCRISPRx-Sth1Cas9-L5 plasmid encoding two sgRNAs will express Sth1Cas9 and dual sgRNAs upon treatment with ATc. The sgRNAs will direct the Sth1Cas9 enzymes towards the designed genomic target regions. Target recognition will lead to the induction of double-stranded DNA breaks which can result in the generation of single indels, double indels, or accurate deletion events upon distinct repair events. Extended deletions beyond the 20 bp sgRNA target site give rise to inaccurate deletions. (B) Percentage of phenotypic crtB mutants identified as white colonies of untreated or ATc-induced M. marinum M using sgRNA-crtb-1 and sgRNA-crtb-2 simultaneously. The percentage phenotypic crtB mutants was calculated as the percentage of white colonies in proportion to the total number of colonies. The mean and standard deviation is shown from two replicate experiments. (C) Percentage of indel and deletion events using dual sgRNAs. White colonies obtained with or without ATc treatment were subjected to PCR and sequencing analysis. Percentages were calculated from the total sequenced colonies per condition (untreated n=19, 1h ATc n=16, 10 days ATc n=13).
4. Discussion
In this study, we provide a novel, fast and efficient single plasmid system to facilitate CRISPR-Cas9-induced mutations in M. marinum and M. tuberculosis. Using our method, we rapidly generated frameshift mutants and precise gene deletions. Previously, the generation of targeted mutants in M. marinum and M. tuberculosis with mycobacterial phages was laborious and time consuming [31]. The recently described oligonucleotide-mediated recombineering followed by Bxb1 integrase targeting (ORBIT) tool enabled efficient genome engineering in mycobacterial species [32]. However, both methods require the insertion of an antibiotic resistance cassette including promoter at the target site which can give rise to polar effects. Removal of these cassettes is time-consuming and leaves scars, which may hinder the generation of additional mutants. Genomic engineering using CRISPR-Cas9 technology circumvents polar effects through the targeted generation of indels resulting in frameshift mutants. Since the discovery of the CRISPR technology, the efficient genome editing tool has been extensively and widely used in many different organisms, including vertebrates, invertebrates, plants and bacteria [33–39]. Recently, CRISPR-mediated gene editing has been established in mycobacteria, where mutants were generated following the introduction of additional NHEJ proteins and inhibition of RecA-dependent repair [10]. On the other hand, our methodology has consistently and rapidly produced frameshift mutations in 86% (25/29) and 100% (7/7) of the genes targeted in M. marinum and M. tuberculosis, respectively (Supplementary Table 4), without overexpression of the NHEJ machinery. We expect that frameshift mutations can be acquired for all genes targeted through the examination of more transformants since only a limited number of transformants was tested. Nevertheless, this demonstrates that the endogenously expressed NHEJ repair mechanism is sufficient to repair CRISPR-Cas9-induced double-stranded DNA breaks in these mycobacteria. DNA repair pathways are highly necessary for bacterial survival as mycobacteria can be exposed to various DNA damaging environments, such as UV radiation, desiccation, antibiotic treatment and host-induced immune responses during infection as reviewed by Gorna et al. [40]. The mycobacterial NHEJ repair system is specifically required in the stationary growth phase [4, 41] to repair double-stranded DNA breaks in the absence of a homologous DNA template, resulting in non-templated addition or deletion of nucleotides [3], and allows gene editing using our single plasmid system. As expected, upon the introduction of a CRISPR-Cas9 plasmid containing the Sth1Cas9 endonuclease and specific sgRNA, a decrease in the number of transformants was detected in comparison to a non-targeting sgRNA in M. marinum, indicating a reduced survival upon induction of double-stranded DNA breaks. Previously, Sun et al. observed that the constitutive expression of several Cas9 proteins resulted in a very low number of transformants in M. smegmatis [9]. Moreover, Yan et al. described that leaky expression of Sth1Cas9 under the control of an ATc-inducible promoter in combination with a targeting sgRNA appeared toxic without overexpression of the NHEJ machinery in M. tuberculosis [10]. CRISPR-Cas12a-mediated genome editing, an alternative to CRISPR-Cas9, has also been successfully applied in M. smegmatis, and in combination with assistance of the NHEJ machinery also in M. marinum [9, 10]. However, CRISPR-Cas12a mediated genome editing in M. marinum was challenging due to the low number of transformants in the presence of a targeting sgRNA, while in M. tuberculosis Cas12a appears almost inactive [10]. Our pCRISPRx-Sth1Cas9-L5 plasmid contains a stronger promoter in front of the Tet Repressor gene than the one used by Yan et al., probably leading to less leaky expression of the CRISPR-Cas9 system, negating the requirement for the expression of the NHEJ machinery. Furthermore, Yan et al. mostly used multicopy plasmids, while we use an integrating plasmid, further reducing the amount of Sth1Cas9 present in the cell. Our results indicate that reduced Sth1Cas9 copy numbers and Sth1Cas9 expression under the control of a strongly repressed anhydrotetracycline (ATc)-inducible promoter yields a sufficient amount of transformants to rapidly induce mutations by the endogenously expressed NHEJ DNA repair proteins in both M. marinum and M. tuberculosis.
In this study, an overall frameshift editing efficiency of 55% and 52% was accomplished in M. marinum and M. tuberculosis, respectively. We speculate that this efficiency is an underestimation as the identification of mutants in our method largely depended on the amplification of the genomic target region and sequencing. Therefore, particularly large deletion events could have been overlooked. We argue, however, that those mutants are undesired as upstream or downstream genes could be affected.
In addition, we observed that for CrtB, both in-frame and out-of-frame deletions abolished protein function. However, frameshift mutations are preferred to ensure disruption of gene function since in-frame mutations will not always result in loss of function.
Using our method, frameshift mutants and gene deletions were successfully achieved following both short and long induction of the CRISPR-Cas9 machinery. Following short induction, however, a higher percentage of accurate deletions was observed, whereas an increased percentage of extended deletions was detected following prolonged ATc induction. In addition, following the extended duration of ATc treatment, an increasing number of PCR products could not be generated for phenotypic crtB mutants acquired with dual sgRNAs (Supplementary Table 5), indicating the possible occurrence of even larger deletions. Our dual sgRNA targeting system also yielded two separate indels simultaneously at distinct target sites within the crtB gene. Consequently, this finding suggests that the presence of dual sgRNAs offers an interesting possibility to simultaneously target at least two distinct genes and thus produce a double mutant in one single step, thereby greatly accelerating mutant production in these slow-growing bacteria. Depending on the experimental aim, the most suitable induction method could be applied to efficiently generate the desired gene deletions or multiple frameshift mutants.
Furthermore, we demonstrated that a gene of interest can be edited distal to the consensus PAM site NNAGAAW, as well as the non-consensus PAM sites NNGGAA and NNAGCAT, thereby greatly expanding the total number of potential target sites in mycobacterial genomes. Sth1Cas9 recognizes an extended PAM sequence in comparison to Streptococcus pyogenes Cas9 (SpCas9), which was suggested to result in a lower number of off-target effects [42]. In agreement with this suggestion, not a single undesired off-target indel was observed in the two M. marinum mutants following short and long exposure to Sth1Cas9 and sgRNA, as was demonstrated by whole genome sequencing. In agreement with our observation, Yan et al detected no (or at most one) off-target mutations when utilizing Sth1Cas9 in M. tuberculosis [10].
Finally, we have shown that the chromosomally integrated CRISPR-Cas9 plasmid can be replaced in a second transformation step with another integrative plasmid to remove the CRISPR-Cas9 machinery. Alternatively, although not explored in this study, replacement of one CRISPR-Cas9 plasmid with a second integrative plasmid containing a CRISPR-Cas9 system targeting another gene could allow for repeated rounds of sequential editing.
Taken together, the adaptation of the previously described CRISPRi technology allowed the efficient production of mutants and targeted genomic deletions in pathogenic mycobacteria. This method enables the possibility to rapidly genetically modify mycobacterial species in one step, which will accelerate research on bacterial pathogenesis and novel strategies to prevent and treat M. tuberculosis infection.
Supplementary Material
(A) M. marinum M WT colonies (yellow) and CRISPR-Cas9-generated non-functional crtB mutant colonies (white) following ATc induction. (B) Efficient replacement of the genomically integrated CRISPR-Cas9 system using the pTdTomato-L5 plasmid in all colonies tested by visualization of tdTomato expression. (C) Isoniazid (INH)-resistance by M. tuberculosis H37Rv mc26020 katG frameshift mutants. Two WT and two katG frameshift mutant strains were spotted on 7H10 plates containing different concentrations of INH and incubated at 37°C for 3 weeks. Colony density was determined by ImageJ analysis.
Acknowledgements
We thank Tessa Montague for adding the mycobacterial genomes to the web interface of CHOPCHOP, Jeremy Rock for questions related to the CRISPR interference system, and Beatriz Izquierdo Lafuente and Jobana Ananthasabesan for providing data of CRISPR-Cas9-generated mutants.
This work was supported by The Cancer Center Amsterdam (to A.S.M.), the ERC Advanced grant ((832721) to S.N), and the Amsterdam Infection and Immunity (AI&II (to A.S.)).
Footnotes
Conflicts of interest
None
References
- [1].WHO. Global Tuberculosis Report. 2019 [Google Scholar]
- [2].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- [4].Stephanou NC, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman MS. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J Bacteriol. 2007;189:5237–5246. doi: 10.1128/JB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d’Adda di Fagagna F, Devine KM, et al. Identification of a DNA Nonhomologous End-Joining Complex in Bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- [6].Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. Mycobacterial Ku and Ligase Proteins Constitute a TwoComponent NHEJ Repair Machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- [7].Doherty AJ, Jackson SP, Weller GR. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Letters. 2001;500:186–188. doi: 10.1016/s0014-5793(01)02589-3. [DOI] [PubMed] [Google Scholar]
- [8].Zheng X, Li S-Y, Zhao G-P, Wang J. An efficient system for deletion of large DNA fragments in Escherichia coli via introduction of both Cas9 and the non-homologous end joining system from Mycobacterium smegmatis. Biochemical and Biophysical Research Communications. 2017;485:768–774. doi: 10.1016/j.bbrc.2017.02.129. [DOI] [PubMed] [Google Scholar]
- [9].Sun B, Yang J, Yang S, Ye RD, Chen D, Jiang Y. A CRISPR-Cpf1-Assisted Non-Homologous End Joining Genome Editing System of Mycobacterium smegmatis. Biotechnol J. 2018;13:e1700588. doi: 10.1002/biot.201700588. [DOI] [PubMed] [Google Scholar]
- [10].Yan MY, Li SS, Ding XY, Guo XP, Jin Q, Sun YC. A CRISPR-Assisted Nonhomologous End-Joining Strategy for Efficient Genome Editing in Mycobacterium tuberculosis. mBio. 2020;11 doi: 10.1128/mBio.02364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, Pritchard JR, Church GM, Rubin EJ, Sassetti CM, Schnappinger D, et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol. 2017;2:16274. doi: 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- [15].Ramakrishnan L, Falkow S. Mycobacterium marinum Persists in Cultured Mammalian Cells in a Temperature-Restricted Fashion. Infect Immun. 1994;62:3222–3229. doi: 10.1128/iai.62.8.3222-3229.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Puttinaowarat S, Thompson K, Lilley J, Adams A. Characterization of Mycobacterium spp. isolated from fish by pyrolysis mass spectrometry (PyMS) analysis. AAHRI Newsl. 1999;8:4–8. [Google Scholar]
- [17].Valway SE, Sanchez MP, Shinnick TF, Orme I, Agerton T, Hoy D, Jones JS, Westmoreland H, Onorato IM. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis . N Engl J Med. 1998;338:633–639. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]
- [18].Steenken W, Oatway WH, Petroff SA. Biological studies of the tubercle bacillus. J Exp Med. 1934;60:515–540. doi: 10.1084/jem.60.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sambandamurthy VK, Derrick SC, Jalapathy KV, Chen B, Russell RG, Morris SL, Jacobs WR., Jr Long-term protection against tuberculosis following vaccination with a severely attenuated double lysine and pantothenate auxotroph of Mycobacterium tuberculosis. Infect Immun. 2005;73:1196–1203. doi: 10.1128/IAI.73.2.1196-1203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sambandamurthy VK, Derrick SC, Hsu T, Chen B, Larsen MH, Jalapathy KV, Chen M, Kim J, Porcelli SA, Chan J, Morris SL, et al. Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine. 2006;24:6309–6320. doi: 10.1016/j.vaccine.2006.05.097. [DOI] [PubMed] [Google Scholar]
- [21].Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ofer N, Wishkautzan M, Meijler M, Wang Y, Speer A, Niederweis M, Gur E. Ectoine biosynthesis in Mycobacterium smegmatis. Appl Environ Microbiol. 2012;78:7483–7486. doi: 10.1128/AEM.01318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].van Winden VJ, Ummels R, Piersma SR, Jimenez CR, Korotkov KV, Bitter W, Houben EN. Mycosins Are Required for the Stabilization of the ESX-1 and ESX-5 Type VII Secretion Membrane Complexes. mBio. 2016;7 doi: 10.1128/mBio.01471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ramakrishnan L, Tran HT, Federspiel NA, Falkow S. A crtB Homolog Essential for Photochromogenicity in Mycobacterium marinum: Isolation, Characterization, and Gene Disruption via Homologous Recombination. J Bacteriol. 1997;179:5862–5868. doi: 10.1128/jb.179.18.5862-5868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Karvelis T, Gasiunas G, Young J, Bigelyte G, Silanskas A, Cigan M, Siksnys V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:253. doi: 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Burggraaf MJ, Speer A, Meijers AS, Ummels R, van der Sar AM, Korotkov KV, Bitter W, Kuijl C. Type VII Secretion Substrates of Pathogenic Mycobacteria Are Processed by a Surface Protease. mBio. 2019;10:e01951-01919. doi: 10.1186/1471-2164-10-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ummels R, Abdallah AM, Kuiper V, Aajoud A, Sparrius M, Naeem R, Spaink HP, van Soolingen D, Pain A, Bitter W. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio. 2014;5:e01744-01714. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang Y, Heymt B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- [31].Chhotaray C, Tan Y, Mugweru J, Islam MM, Adnan Hameed HM, Wang S, Lu Z, Wang C, Li X, Tan S, Liu J, et al. Advances in the development of molecular genetic tools for Mycobacterium tuberculosis. J Genet Genomics. 2018;45:281–297. doi: 10.1016/j.jgg.2018.06.003. [DOI] [PubMed] [Google Scholar]
- [32].Murphy KC, Nelson SJ, Nambi S, Papavinasasundaram K, Baer CE, Sassetti CM. ORBIT: a New Paradigm for Genetic Engineering of Mycobacterial Chromosomes. mBio. 2018;9 doi: 10.1128/mBio.01467-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Oh JH, van Pijkeren JP. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014;42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cong L, RF A, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;338:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mali P, Yang L, Esvelt KM, Aach J, Guell M, Di Carlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- [39].Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi J, Qiu J, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- [40].Gorna AE, Bowater RP, Dziadek J. DNA repair systems and the pathogenesis of Mycobacterium tuberculosis: varying activities at different stages of infection. Clin Sci (Lond) 2010;119:187–202. doi: 10.1042/CS20100041. [DOI] [PubMed] [Google Scholar]
- [41].Pitcher RS, Green AJ, Brzostek A, Korycka-Machala M, Dziadek J, Doherty AJ. NHEJ protects mycobacteria in stationary phase against the harmful effects of desiccation. DNA Repair (Amst) 2007;6:1271–1276. doi: 10.1016/j.dnarep.2007.02.009. [DOI] [PubMed] [Google Scholar]
- [42].Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24:636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) M. marinum M WT colonies (yellow) and CRISPR-Cas9-generated non-functional crtB mutant colonies (white) following ATc induction. (B) Efficient replacement of the genomically integrated CRISPR-Cas9 system using the pTdTomato-L5 plasmid in all colonies tested by visualization of tdTomato expression. (C) Isoniazid (INH)-resistance by M. tuberculosis H37Rv mc26020 katG frameshift mutants. Two WT and two katG frameshift mutant strains were spotted on 7H10 plates containing different concentrations of INH and incubated at 37°C for 3 weeks. Colony density was determined by ImageJ analysis.