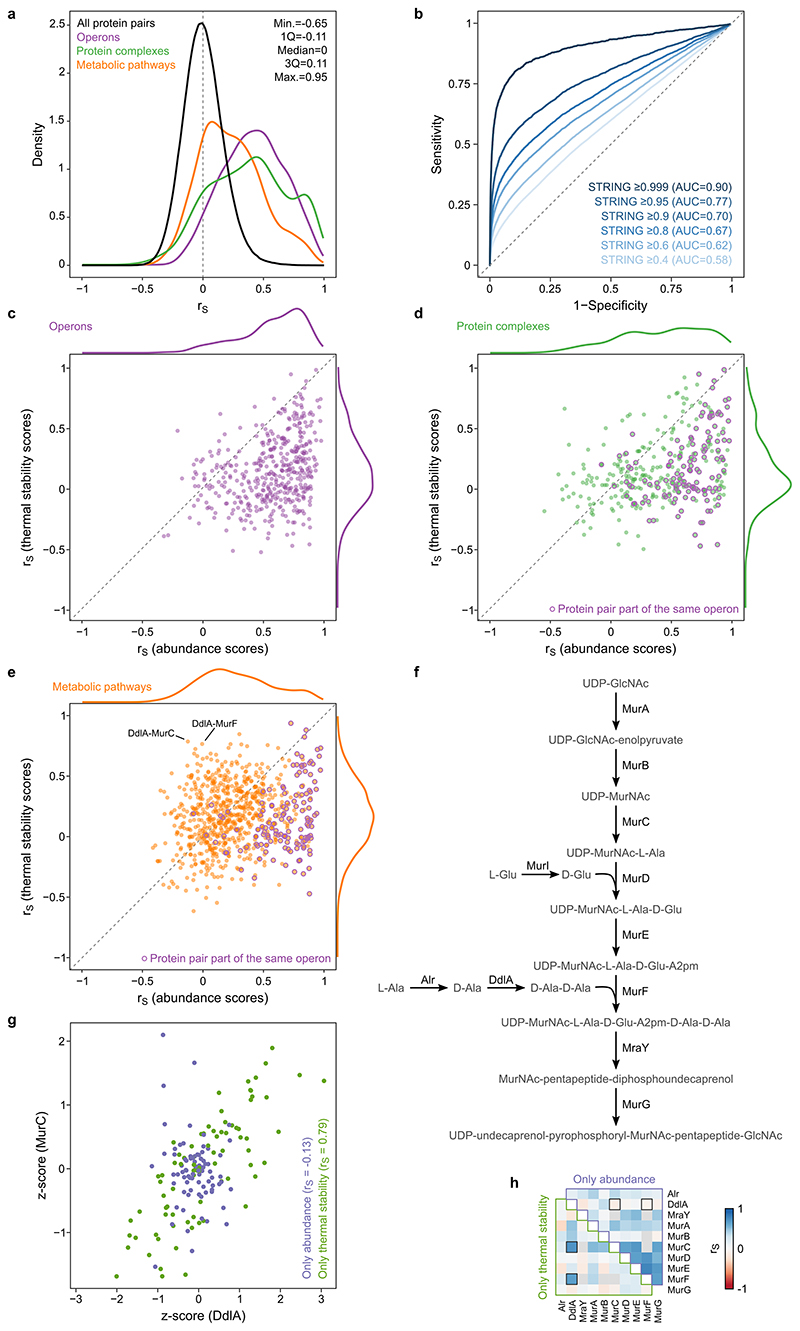
Extended Data Figure 6. Protein correlation profiling recapitulates known biological interactions with abundance and thermal stability data having different contribution to functional associations.
(a) Distribution of Spearman’s rank correlation of all protein pair comparisons compared to known operons, protein complexes, and metabolic pathways. Distribution statistics refer to all protein pairs. (b) ROC analysis based on the decreasing absolute Spearman’s rank correlation compared to interactions in STRING database at different cut-offs of the combined STRING score. (c-e) Spearman’s rank correlation of protein pairs belonging to the same operon (c), protein complex (d), or metabolic pathway (e) using solely abundance changes (x-axis) or thermal stability changes (y-axis). Protein pairs belonging to the same operon are highlighted in purple. Distribution of Spearman’s rank correlation are shown outside the axes. n=446 for operons, n=348 for protein complexes, and n=801 for metabolic pathways. Proteins belonging to the same operon or complex mostly have coordinated abundance changes, while proteins belonging to the same pathway have also often coordinated thermal stability. (f) Schematic representation of UDP-N-acetylmuramoyl-pentapeptide biosynthesis pathway. (g) Example of protein pair (DdlA and MurC) co-changing in their thermal stability (rS=0.79), but not abundance (rS=-0.13) across 81 genetic perturbations. Each data point corresponds to the abundance or thermal stability z-score in one of the genetic perturbations (color coded). (h) Heatmap of Spearman’s rank correlation of all quantified members of UDP-N-acetylmuramoyl-pentapeptide biosynthesis pathway based on co-changes in abundance (upper triangle) or thermal stability alone (lower triangle).